Neurovascular Coupling in fMRI and fNIRS: Mechanisms, Methods, and Clinical Translation for Biomedical Research
This article provides a comprehensive exploration of neurovascular coupling (NVC) as the fundamental physiological principle underlying functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS).
Neurovascular Coupling in fMRI and fNIRS: Mechanisms, Methods, and Clinical Translation for Biomedical Research
Abstract
This article provides a comprehensive exploration of neurovascular coupling (NVC) as the fundamental physiological principle underlying functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS). Tailored for researchers, scientists, and drug development professionals, it details the cellular mechanisms of NVC, compares the methodological applications and signal origins of fMRI and fNIRS, discusses troubleshooting and optimization strategies for data interpretation, and validates findings through multi-modal integration. The review also examines the critical role of NVC dysfunction in neurodegenerative and cerebrovascular diseases, highlighting its potential as a biomarker for diagnosis and therapeutic intervention in clinical trials and preclinical research.
The Biological Basis of Neurovascular Coupling: From Cellular Mechanisms to Functional Imaging Signals
Defining Neurovascular Coupling and the Neurovascular Unit (NVU)
Neurovascular coupling (NVC) is the fundamental physiological mechanism that links transient neural activity to corresponding, localized changes in cerebral blood flow (CBF) [1] [2]. This process, also termed functional hyperemia, ensures that activated brain regions receive an immediate and precise supply of oxygen and glucose to meet elevated metabolic demands [2]. The brain, despite accounting for only about 2% of total body weight, consumes approximately 20% of the body's total energy, yet possesses minimal energy reserves, making this tight coupling critical for normal function [3] [4]. The concept was first articulated by Roy and Sherrington in 1890, forming the Roy-Sherrington principle which states that the brain possesses an intrinsic mechanism to vary its vascular supply locally in correspondence with local variations of functional activity [2].
The functional complex that executes this process is the neurovascular unit (NVU). The NVU is an integrated system comprising neurons, astrocytes, and vascular cells (including endothelial cells, pericytes, and vascular smooth muscle cells), which work in concert to regulate cerebral blood flow [5] [6] [2]. The formal concept of the NVU was introduced by the National Institute of Neurological Disorders and Stroke (NINDS) in 2001, highlighting the interconnected relationship between brain cells and blood vessels that was previously underappreciated [7] [6]. The NVC process orchestrated by the NVU forms the physiological basis for functional neuroimaging techniques such as functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS), which measure hemodynamic changes as proxies for neural activity [1] [2].
Anatomical Composition of the Neurovascular Unit
The NVU is a multi-cellular structure that facilitates communication between neural tissue and the cerebral vasculature. Its components exist along a three-dimensional network of pial and penetrating arterioles, capillaries, venules, and veins [5]. The radial composition of the capillary NVU, where the strongest blood-brain barrier properties are manifest, includes:
- Endothelial Cells: A continuous monolayer of specialized endothelial cells lines the brain's blood vessels, forming a physical barrier through circumferentially interconnected transmembrane tight junction proteins (e.g., claudins, occludin) [5]. These cells exhibit unique properties compared to peripheral endothelial cells, including suppressed transcytosis and polarized expression of various transporters, which together create a highly selective interface between blood and brain [5].
- Basement Membrane: A non-cellular, compact lattice meshwork composed of proteins including collagens, laminins, nidogens, perlecan, and agrin [5]. This membrane stabilizes the blood-brain barrier, anchors cells, facilitates signal transduction, and is dynamically maintained by bidirectional signaling with other NVU components [5].
- Mural Cells: This category includes pericytes on capillaries and vascular smooth muscle cells (VSMCs) on arterioles. Capillaries in the central nervous system have the highest pericyte coverage in the body (pericyte to endothelial cell ratio of approximately 1:4) [5]. Pericytes are embedded between the endothelial tube and astrocyte endfeet and play critical roles in BBB establishment, vascular development, and regulation of capillary blood flow [5]. VSMCs control vessel diameter through contraction and relaxation.
- Astrocytes: These glial cells project endfeet processes that completely ensheathe the vascular components, while their fine processes contact neuronal synapses [5]. This unique positioning allows astrocytes to transmit signals from neurons to the vasculature [5].
- Microglia: The brain's resident immune cells are also present within the NVU interface and contribute to neuroimmune responses [5].
- Neurons: Various neuronal subtypes, including pyramidal neurons and interneurons, initiate the NVC response through the release of neurotransmitters and vasoactive substances [3].
Table 1: Cellular Components of the Neurovascular Unit
| Cell Type | Primary Location | Key Functions in NVU |
|---|---|---|
| Endothelial Cells | Lumen of blood vessels | Form the physical barrier; regulate transport; release signaling molecules [5]. |
| Pericytes | Capillary wall (embedded in basement membrane) | Regulate capillary diameter; contribute to BBB integrity; produce extracellular matrix [5]. |
| Vascular Smooth Muscle Cells | Arteriole wall | Control arteriolar diameter; responsible for bulk blood flow regulation [1]. |
| Astrocytes | Parenchyma, with endfeet on vessels | Transmit signals from neurons to vasculature; release vasoactive substances [5]. |
| Neurons | Parenchyma | Initiate NVC via neurotransmitter release; different subtypes contribute specific vasoactive signals [3]. |
| Microglia | Parenchyma | Neuroimmune surveillance; can influence vascular function and integrity [5]. |
Molecular Mechanisms of Neurovascular Coupling
NVC is mediated by complex cellular signaling pathways that are initiated by synaptic activity and culminate in vascular dilation. The process involves a coordinated sequence of events across different NVU components, as illustrated in the following diagram:
NVC signaling involves multiple parallel pathways that are temporally coordinated [3] [4]:
Neuronal Signaling: Neurons directly influence blood vessels through the release of vasoactive substances. Glutamate-mediated activation of neurons leads to the release of potent vasodilators including nitric oxide (NO) from nitrergic interneurons and prostaglandin E2 (PGE2) from pyramidal neurons [1] [4]. Quantitative modeling of optogenetics data in mice has revealed that different neuronal sub-populations contribute to distinct temporal phases of the vascular response: the first rapid dilation is caused by NO-interneurons, the main sustained dilation during longer stimuli is caused by pyramidal neurons, and the post-stimulus undershoot is regulated by NPY-interneurons [3].
Astrocytic Signaling: Astrocytes transmit signals from synapses to blood vessels primarily through calcium (Ca²⁺) signaling [2]. Increased Ca²⁺ in astrocytes triggers the release of various vasoactive substances including potassium ions (K⁺) through large-conductance calcium-activated potassium (BK) channels and inward rectifier potassium (Kir) channels on astrocytic endfeet, as well as metabolites of arachidonic acid such as prostaglandins and epoxyeicosatrienoic acids (EETs) [1] [2]. The vasomotor effect of astrocytic signaling is bidirectional, with moderate Ca²⁺ increases inducing vasodilation and larger increases potentially causing vasoconstriction [2].
Vascular Response: The released vasoactive molecules act on smooth muscle cells of arterioles and pericytes of capillaries to induce relaxation and vessel dilation [1]. This dilation starts locally and back-propagates through endothelial cell signaling via gap junctions along the vascular tree to reach larger arteries, coordinating a regional hemodynamic response [1] [5]. While traditionally thought to be primarily controlled by arteriolar smooth muscle cells, recent evidence suggests that capillary pericytes may also participate in vasodilation during brain activation, potentially acting even faster than smooth muscle cells [1].
Research Methodologies and Experimental Protocols
In Vivo Human Assessment Techniques
Non-invasive neuroimaging techniques form the cornerstone of human NVC research, leveraging the coupling between neural activity and hemodynamics to infer brain function.
Functional Magnetic Resonance Imaging (fMRI): The most widely used technique, fMRI typically measures the blood oxygenation level-dependent (BOLD) signal [1] [2]. The BOLD signal reflects changes in the ratio of oxygenated to deoxygenated hemoglobin following neural activity-induced changes in CBF and oxygen metabolism [1]. The typical hemodynamic response function (HRF) to a brief stimulus consists of a slight initial dip (debated), a main peak at approximately 3-6 seconds, and a post-stimulus undershoot before returning to baseline [3] [2]. Arterial Spin Labeling (ASL) is another fMRI technique that directly quantifies CBF by magnetically labeling arterial blood water as an endogenous tracer, though it has lower temporal resolution than BOLD [1].
Functional Near-Infrared Spectroscopy (fNIRS): This optical neuroimaging technique measures relative changes in cerebral oxygenation by detecting light attenuation at different wavelengths as it passes through brain tissue [8]. fNIRS provides measures of oxyhemoglobin (O₂Hb), deoxyhemoglobin (HHb), and total hemoglobin (tHb), with tHb serving as a proxy for cerebral blood volume [8]. While fNIRS has poorer spatial resolution and cannot access deep brain structures compared to fMRI, it offers higher temporal resolution (can reach 1 ms), is more portable, and less susceptible to movement artifacts [1] [8].
A representative experimental protocol for assessing NVC using fNIRS is outlined below, adapted from research on sport-related concussion in retired athletes [8]:
Table 2: Experimental Protocol for fNIRS-based NVC Assessment
| Protocol Phase | Duration | Procedure | Measured Variables |
|---|---|---|---|
| Baseline Recording | 5 minutes | Participant sits quietly, eyes open, breathing normally, no communication. | Baseline O₂Hb, HHb, and tHb concentrations. |
| NVC Task (Where's Wally) | 5 cycles (5 minutes total) | Each cycle: 20s eyes closed, 40s eyes open searching for "Wally" in an image. If found early, advance to next image. | Task-induced changes in O₂Hb, HHb, and tHb. Calculation of HbDiff (O₂Hb - HHb) to assess oxygen extraction. |
| Data Analysis | - | Compare hemodynamic response patterns between groups (e.g., patients vs. controls). | Amplitude, timing, and morphology of hemodynamic responses. |
Multimodal Imaging and Computational Modeling
Advanced research approaches combine multiple techniques to gain a more comprehensive understanding of NVC:
Multimodal Integration: Simultaneous measurement of electrophysiological (e.g., local field potentials - LFP) and hemodynamic signals provides direct insight into neurovascular relationships [3] [4]. For example, simultaneous LFP and fMRI measurements in primates have demonstrated that the BOLD signal correlates more closely with synaptic activity (LFPs) than with spiking output [4] [2].
Computational Modeling: Quantitative mathematical models integrate data from multiple species and experimental modalities to create unified frameworks of NVC [3]. A recent comprehensive model combines mechanistic understanding of cellular signaling pathways (from optogenetics in mice) with Windkessel models of blood flow dynamics and the biophysics of the BOLD signal [3] [4]. Such models can predict the contributions of specific neuronal sub-populations to different phases of the hemodynamic response and facilitate the translation of insights from animal studies to human applications [3].
Quantitative Parameters and Hemodynamic Response
The hemodynamic response measured by fMRI and fNIRS follows characteristic temporal dynamics and can be quantified using specific parameters. The table below summarizes key quantitative metrics derived from NVC research:
Table 3: Quantitative Parameters in Neurovascular Coupling Research
| Parameter | Typical Values / Characteristics | Biological Significance | Measurement Techniques |
|---|---|---|---|
| Hemodynamic Response Function (HRF) Timing | Peak: 3-6 s post-stimulus; Duration: 15-20 s [3] | Reflects speed and duration of CBF response to neural activity. | BOLD-fMRI, fNIRS |
| CBF Increase During Activation | 20-40% above baseline [1] | Magnitude of functional hyperemia; indicates vascular reactivity. | ASL, Laser Doppler/Speckle Flowmetry |
| Baseline CBF | Accepts ~20% of cardiac output [3] [9] | Reflects resting state metabolic support. | ASL, PET |
| BOLD Signal Change | Typically 0.5-5% at 3T [2] | Indirect measure of the balance between CBF and CMRO₂. | BOLD-fMRI |
| Cell-Specific Response Contributions | NO-interneurons: rapid dilation; Pyramidal neurons: sustained dilation; NPY-interneurons: post-stimulus undershoot [3] | Links specific neural elements to vascular response phases. | Optogenetics with microscopy (animal models) |
| Spatial Specificity | Vascular response localized to activated cortical columns [1] | Precision of metabolic supply to active neural tissue. | High-resolution fMRI, optical imaging |
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 4: Key Research Reagent Solutions for Neurovascular Coupling Studies
| Reagent / Material | Category | Primary Function in NVC Research |
|---|---|---|
| Optogenetic Constructs (e.g., Channelrhodopsin) | Genetic Tool | Selective activation of specific neuronal sub-populations (e.g., pyramidal cells, NO- or NPY-interneurons) to dissect their contribution to hemodynamic responses [3]. |
| Vasoactive Compound Inhibitors (e.g., L-NAME for NOS, COX inhibitors) | Pharmacological Agent | Selective blockade of specific signaling pathways (e.g., NO synthesis, prostaglandin production) to determine their role in functional hyperemia [1] [2]. |
| Calcium Indicators (e.g., GCaMP) | Imaging Probe | Monitoring intracellular Ca²⁺ dynamics in astrocytes and neurons during neural activation, a key signaling event in NVC [2]. |
| Human-derived Brain Cells (HBMVEC, HBVP, HA, HM, HO, HN) | Cell Culture Model | Creating physiologically relevant in vitro human NVU models for drug screening and disease modeling, overcoming species-specific limitations [10]. |
| Arterial Spin Labeling (ASL) MRI Sequences | Imaging Sequence | Non-invasive quantification of cerebral blood flow (CBF) changes during neural activation, providing a direct hemodynamic metric [1] [9]. |
| fNIRS Systems (e.g., multi-channel systems) | Imaging Hardware | Portable, high temporal resolution monitoring of cortical oxygenation changes (O₂Hb, HHb) during cognitive or sensory tasks [8]. |
Clinical Implications and Pathophysiological Significance
NVC dysfunction, often termed neurovascular uncoupling, is implicated in a wide range of neurological and psychiatric disorders, making it a significant focus for therapeutic development.
Neurodegenerative Diseases: In Alzheimer's disease, impaired functional hyperemia has been detected both in animal models and humans, often before the appearance of amyloid plaques [1] [7]. Multiple mechanisms contribute, including amyloid-β-induced endothelial dysfunction, pericyte loss, and oxidative stress, which disrupt the NVU's ability to match blood flow to neural demand [7] [2]. Similar NVC alterations occur in Parkinson's disease and Huntington's disease [7].
Cerebrovascular Disorders: Hypertension, small vessel disease, and cerebral amyloid angiopathy can damage arterioles and capillaries, altering the adaptive response of the cerebral microvasculature [1] [7]. After ischemic stroke, NVC impairments can occur remotely from the infarction site due to transhemispheric diaschisis [1].
Psychiatric Disorders: Recent research in major depressive disorder has identified NVC decoupling as a potential neuropathological mechanism [9]. Multimodal MRI studies in drug-naïve MDD patients have revealed reduced spatial correlation between neuronal activity (ALFF) and cerebral blood flow, with distinct patterns based on disease severity and sex [9].
Traumatic Brain Injury: Sport-related concussions and repeated mild traumatic brain injuries can lead to long-term NVC alterations, as evidenced by reduced cerebral hemodynamic responses in retired athletes with a history of multiple head injuries [8]. These changes potentially reflect underlying endothelial dysfunction and impaired cerebrovascular reactivity [8].
Therapeutic strategies aimed at preserving or restoring NVC function include interventions that improve endothelial function, reduce oxidative stress (e.g., inhibition of NADPH oxidases), increase nitric oxide bioavailability, and potentially target specific signaling pathways within the NVU [1] [2]. The development of sophisticated human-derived NVU models is expected to accelerate the discovery of such therapies by providing more physiologically relevant platforms for drug screening and disease modeling [10].
The neurovascular unit (NVU) is an integrated system comprising neurons, astrocytes, and vascular cells that coordinates cerebral blood flow (CBF) with neuronal activity, a process known as neurovascular coupling (NVC) [2]. This physiological mechanism ensures the precise and rapid delivery of oxygen and nutrients to active brain regions, forming the fundamental basis for functional neuroimaging techniques such as fMRI and fNIRS [11] [12]. The cellular players within the NVU—neurons, astrocytes, vascular smooth muscle cells (VSMCs), and pericytes—orchestrate complex signaling pathways to achieve this tight regulation. Understanding their distinct roles, interactions, and the experimental methods used to study them is crucial for interpreting neuroimaging data and developing therapies for neurological diseases where NVC is impaired [11] [13].
Table: Core Cellular Components of the Neurovascular Unit
| Cell Type | Primary Location | Key Functions in NVC | Major Vasoactive Signals |
|---|---|---|---|
| Neurons | Brain parenchyma | Initiate vasodilation via neurotransmitter release; pyramidal neurons act as "neurogenic hubs" [11]. | Glutamate, Nitric Oxide (NO), Prostaglandin E2 (PGE2), ATP [11] [2] |
| Astrocytes | Interposed between synapses and vasculature | Connect synaptic activity to vascular response; release vasoactive factors via Ca²⁺ signaling [11] [14]. | Potassium (K⁺), Prostaglandins (PGE₂), Epoxyeicosatrienoic Acids (EETs), Glutamate [13] [2] |
| Vascular Smooth Muscle Cells (VSMCs) | Arterioles and arteries | Regulate large-scale, rapid changes in blood flow by contracting/relaxing to change arteriolar diameter [13] [15]. | Respond to NO, PGE₂, EETs, K⁺; cyclic GMP-mediated relaxation [13] |
| Pericytes | Capillaries (ensheathing, midcapillary, postcapillary subtypes) [15] | Control slower, local capillary tone and flow; involved in initial capillary dilation; maintain BBB [13] [15] [16]. | Respond to glutamate, NO, PGE₂ (via EP4 receptors), Angiotensin II; contract via ATP (P2X7 receptors) [13] [15] |
Neurons: Initiators of Hemodynamic Signals
Mechanisms and Signaling Pathways
Neurons, particularly excitatory pyramidal cells, serve as the primary initiators of NVC, functioning as "neurogenic hubs" [11]. Upon activation, these neurons release neurotransmitters and vasoactive mediators that trigger vasodilation. A key pathway involves glutamate, which acts on both astrocytes and neurons. Crucially, neuronal activity leads to calcium influx, activating neuronal nitric oxide synthase (nNOS) in specific interneurons, resulting in the production of nitric oxide (NO), a potent vasodilator [11] [13]. A systematic review concluded that nNOS blockade causes the most substantial reduction in neurovascular response, averaging 64% across studies [13]. Furthermore, pyramidal neurons have been identified as a major cellular source of prostaglandin E2 (PGE2), which induces vasodilation by acting on EP2 and EP4 receptors on vascular cells [11]. Inhibitory GABAergic interneurons also contribute by modulating the output of pyramidal cells, with some studies showing they can regulate blood flow via NO release [11].
Key Experimental Protocols and Reagents
Studying neuronal contributions to NVC often involves controlled stimulation and targeted inhibition of specific pathways.
- Whisker Stimulation in Rodents: A classic in vivo model where mechanical deflection of the whiskers activates neurons in the barrel cortex. The ensuing changes in local field potential (LFP) and CBF (measured via laser Doppler or laser speckle flowmetry) are recorded [11].
- Optogenetics: Channelrhodopsin-2 (ChR2) is expressed in specific neuronal populations (e.g., pyramidal neurons). Light pulses are delivered via an optical fiber to selectively activate these neurons, allowing researchers to dissect their specific contribution to the hemodynamic response without confounding sensory inputs [11].
- Pharmacological Blockade: The role of specific mediators is tested by applying inhibitors. For example, the nonspecific NOS inhibitor L-NAME or the specific nNOS inhibitor S-Methyl-L-thiocitrulline (SMTC) is used to quantify the contribution of NO to the hyperemic response [13].
Table: Research Reagent Solutions for Neuronal NVC Studies
| Reagent / Tool | Function / Target | Experimental Application |
|---|---|---|
| L-NAME | Non-selective Nitric Oxide Synthase (NOS) inhibitor | Broadly blocks NO production to assess its overall role in NVC [13]. |
| S-Methyl-L-thiocitrulline (SMTC) | Selective neuronal NOS (nNOS) inhibitor | Specifically targets neuronal NO pathways, isolating their contribution [13]. |
| NS-398 | Selective COX-2 inhibitor | Blocks prostaglandin synthesis, used to probe the PGE2 pathway [11]. |
| Channelrhodopsin-2 (ChR2) | Light-gated cation channel for optogenetics | Enables precise, millisecond-timescale activation of specific neuronal populations to evoke NVC responses [11]. |
| Local Field Potential (LFP) Recording | Measures summed synaptic activity from neuronal populations | Serves as a direct electrophysiological correlate of neural activity to compare with hemodynamic changes [11] [14]. |
Astrocytes: Bridges between Synapses and Vasculature
Mechanisms and Signaling Pathways
Astrocytes, with their endfeet ensheathing cerebral blood vessels, are ideally positioned to relay signals from synapses to the vasculature [11]. The traditional view involves glutamate from synaptic activity activating metabotropic glutamate receptors (mGluR5) on astrocytes, triggering intracellular calcium (Ca²⁺) waves. This Ca²⁺ increase leads to the production and release of several vasoactive agents [11] [2]. These include:
- Potassium (K⁺): Efflux through BK channels on astrocytic endfeet into the perivascular space, leading to VSMC hyperpolarization and dilation [2].
- Arachidonic Acid Metabolites: Phospholipase D2 (PLD2)-mediated production of arachidonic acid, which is metabolized by cyclooxygenase-1 (COX1) to the vasodilator PGE2, and by cytochrome P450 enzymes to epoxyeicosatrienoic acids (EETs) [13].
- Glutamate: Astrocytes can also release glutamate, which may act directly on vessels [11].
However, the role of astrocytic Ca²⁺ is debated. Some studies suggest it is necessary for capillary dilation via pericytes but not for arteriolar dilation, and the relevance of mGluR5 in the adult brain has been questioned [13].
Key Experimental Protocols and Reagents
Astrocytic function is probed by inhibiting their metabolism and manipulating key signaling molecules.
- In Vivo Calcium Imaging: Genetically encoded calcium indicators (e.g., GCaMP) are expressed in astrocytes. Two-photon microscopy is then used to visualize stimulus-evoked Ca²⁺ dynamics in astrocytic endfeet in vivo in relation to vessel diameter changes [12].
- Metabolic Inhibition with Fluorocitrate: Fluorocitrate is a reversible inhibitor of the astrocyte-specific Krebs cycle enzyme aconitase. Local administration selectively impairs astrocyte metabolism, allowing researchers to assess its impact on NVC by comparing blood flow responses (e.g., SCBF) before and after application while monitoring neural activity (LFP) [14].
- Genetic Knockout Models: Mice lacking inositol 1,4,5-trisphosphate receptor type 2 (IP3R2-KO) have impaired astrocytic Ca²⁺ signaling. These models are used to study the necessity of astrocytic Ca²⁺ waves in functional hyperemia [13].
Table: Research Reagent Solutions for Astrocytic NVC Studies
| Reagent / Tool | Function / Target | Experimental Application |
|---|---|---|
| Fluorocitrate | Inhibits astrocyte-specific aconitase | Used to reversibly impair astrocyte metabolism and assess its role in NVC without directly blocking neurons [14]. |
| IP3R2-Knockout Mice | Lacks primary Ca²⁺ release mechanism in astrocytes | Genetic model to study the necessity of astrocytic Ca²⁺ signaling in functional hyperemia [13]. |
| mGluR5 Antagonists (e.g., MTEP) | Blocks metabotropic glutamate receptor 5 | Tests the traditional glutamate-induced Ca²⁺ signaling pathway in astrocytes, though its role in adults is limited [13]. |
| Two-Photon Microscopy | High-resolution deep-tissue imaging | Allows real-time visualization of Ca²⁺ dynamics in astrocytic endfeet and simultaneous measurement of capillary/arteriole diameter in vivo [12]. |
Vascular Smooth Muscle Cells and Pericytes: Effectors of Blood Flow
Mechanisms and Signaling Pathways
VSMCs and pericytes are the contractile cells that directly regulate vessel diameter. VSMCs surround arterioles and control large-scale, rapid changes in blood flow, while pericytes, embedded in the capillary basement membrane, control local capillary tone and flow heterogeneity [13] [15]. They respond to vasoactive signals from neurons and astrocytes by relaxing, thereby dilating the vessel.
- Nitric Oxide (NO): NO diffuses into VSMCs and activates soluble guanylate cyclase (sGC), producing cyclic GMP (cGMP), which leads to relaxation. In pericytes, NO may act indirectly by inhibiting the vasoconstrictor 20-HETE [13].
- Prostaglandin E2 (PGE2): PGE2 binds to Gₛ-coupled EP4 receptors on pericytes and VSMCs, increasing intracellular cAMP and promoting relaxation [13].
- Potassium (K⁺): Elevated external K⁺ (5-20 mM) leads to VSMC and pericyte hyperpolarization by activating inward-rectifier K⁺ (Kir) channels, closing voltage-gated calcium channels, and causing relaxation [2].
- Constrictor Signals: ATP can induce pericyte contraction via P2X7 receptors, and high concentrations of PGE2 can cause vasoconstriction via EP1 receptors [13] [15].
Pericytes are heterogeneous; ensheathing pericytes at the arteriole-capillary junction express high levels of α-smooth muscle actin and are primarily responsible for regulating microvascular blood flow, while mid-capillary pericytes control local capillary tone [15].
Key Experimental Protocols and Reagents
Investigating contractile cells involves isolating their responses and visualizing their dynamics.
- Two-Photon Microscopy of Pericyte Ca²⁺ and Diameter: This technique is used in awake, head-fixed mice to image Ca²⁺ fluctuations in pericytes and simultaneously measure the diameter of associated capillaries in response to sensory stimulation or pharmacological application [11] [15].
- Laser Speckle Contrast Imaging (LSCI): A wide-field optical technique that provides high spatiotemporal resolution maps of CBF changes. It is often used in conjunction with stimulation (e.g., whisker, forepaw) to visualize the cortical hemodynamic response and assess the impact of manipulating VSMCs or pericytes [12].
- Isolated Vessel Myography: Parenchymal arterioles are surgically removed and maintained in a bath. Their diameter changes in response to vasoactive drugs (e.g., PGE2, NO donors) applied directly to the bath are measured, allowing for controlled study of VSMC pharmacology [13].
Table: Research Reagent Solutions for Studying Contractile Cells
| Reagent / Tool | Function / Target | Experimental Application |
|---|---|---|
| ODQ | Inhibitor of soluble guanylate cyclase (sGC) | Blocks the NO-cGMP signaling pathway in VSMCs to test its specific role in vasodilation [13]. |
| HET0016 | Selective inhibitor of 20-HETE synthesis | Used to probe the role of the vasoconstrictor 20-HETE in capillary perfusion and its interaction with NO signaling in pericytes [13]. |
| L-NAME | Nitric Oxide Synthase (NOS) inhibitor | Reduces endogenous NO production, leading to impaired vasodilation and used to study NO's role in NVC [13]. |
| PF-04418948 | Selective EP2 receptor antagonist | Pharmacologically blocks the PGE2 EP2 receptor to investigate its role in pericyte-mediated capillary dilation [13]. |
| Two-Photon Microscopy | High-resolution deep-tissue imaging | Enables direct in vivo observation of pericyte Ca²⁺ signals and contractile dynamics alongside capillary diameter changes [15]. |
Integrated Experimental Workflow for NVC Investigation
A comprehensive investigation of NVC in animal models typically integrates multiple techniques to correlate neural activity with hemodynamic responses and pinpoint cellular mechanisms.
Table: Quantitative Data on Key NVC Signaling Pathways
| Signaling Pathway | Primary Mediator | Cellular Source | Vascular Target | Reported Impact on CBF Response |
|---|---|---|---|---|
| Nitric Oxide (NO) | nNOS-derived NO | nNOS Interneurons [11] | VSMCs, Pericytes [13] | ~64% reduction with nNOS blockade [13] |
| Prostaglandins | PGE2 | Pyramidal Neurons, Astrocytes [11] | Pericyte EP4 Receptors [13] | Significant component; EP4 blockade inhibits dilation [13] |
| Potassium (K⁺) | K⁺ (5-20 mM) | Astrocytic Endfeet (BK/Kir channels) [2] | VSMCs, Pericytes [2] | Key mechanism for astrocyte-mediated vasodilation [2] |
| EETs | Epoxyeicosatrienoic Acids | Astrocytes (CYP450) [13] | Capillary Pericytes [13] | Contributes to capillary-level NVC [13] |
| Astrocytic Ca²⁺ | Ca²⁺ waves | Astrocytes [11] | Various downstream effectors | Necessary for capillary but not arteriolar dilation in some studies [13] |
Neurovascular coupling (NVC) is the fundamental biological process that ensures a rapid and precise increase in cerebral blood flow (CBF) to regions of heightened neural activity, a mechanism also known as functional hyperemia [17] [11]. The brain's impressive energy demands, coupled with its very limited energy reserves, make this dynamic regulation of blood supply critically important for sustaining normal neural function [17]. The coordinated interaction of vasoactive signaling molecules—primarily glutamate, nitric oxide (NO), prostaglandins (PGs), and potassium (K+)—ensures that active neurons receive adequate oxygen and nutrients [1] [11]. This process is orchestrated by the neurovascular unit (NVU), a complex network comprising neurons, astrocytes, vascular smooth muscle cells (VSMCs), and pericytes [11].
Understanding these signaling pathways is not merely an academic exercise; it is essential for interpreting functional neuroimaging data. Techniques such as functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS) rely on hemodynamic signals that are ultimately governed by these molecular players [1] [18]. Furthermore, the failure of NVC is increasingly recognized as an early event in various cerebrovascular and neurodegenerative diseases, making these pathways promising targets for therapeutic intervention [17] [11]. This whitepaper provides an in-depth technical guide to the roles, interactions, and experimental investigation of these key vasoactive signals.
Detailed Mechanisms of Key Vasoactive Signals
Glutamate: The Primary Initiator
As the main excitatory neurotransmitter in the brain, glutamate serves as the canonical trigger for the NVC response [17]. Its actions are mediated through two distinct receptor classes located on different cell types:
- Ionotropic Glutamate Receptors (iGluRs) on Neurons: The activation of N-methyl-D-aspartate receptors (NMDAr) on postsynaptic neurons is a central pathway. Glutamate binding and subsequent Ca²⁺ influx through NMDAr lead to the activation of neuronal nitric oxide synthase (nNOS) and the production of nitric oxide (NO), a potent vasodilator [17]. The enzyme nNOS is strategically anchored to the synaptic membrane via a supramolecular complex with NMDAr and the scaffold protein PSD-95, ensuring rapid and localized NO production in response to glutamate release [17].
- Metabotropic Glutamate Receptors (mGluRs) on Astrocytes: Glutamate also activates G-protein coupled mGluRs on astrocytes, initiating a calcium signaling cascade within these glial cells. This results in the synthesis and release of various vasoactive metabolites, including epoxyeicosatrienoic acids (EETs) and prostaglandins (PGs), which act on adjacent arterioles [1] [11].
Table 1: Glutamate Receptors in Neurovascular Coupling
| Receptor Type | Primary Location | Signaling Pathway | Key Vasoactive Output |
|---|---|---|---|
| NMDA Receptor (iGluR) | Postsynaptic Neurons | Ca²⁺ influx → nNOS activation | Nitric Oxide (NO) |
| Metabotropic Receptor (mGluR) | Astrocytes | G-protein → Ca²⁺ release → PLA₂/COX activation | Prostaglandins, EETs |
Nitric Oxide (NO): The Gaseous Messenger
Nitric oxide is a ubiquitous, gaseous signaling molecule recognized as a key player in NVC, essential for the full development of the neurovascular response [17]. Its synthesis, bioavailability, and signaling are tightly regulated through multiple pathways.
- Synthesis via NOS Enzymes: The classical pathway for •NO production is the enzymatic conversion of L-arginine to L-citrulline by a family of nitric oxide synthases (NOS), requiring oxygen and several cofactors (NADPH, FAD, FMN, heme, BH₄) [17] [19]. Three major isoforms exist:
- Neuronal NOS (nNOS): Constitutively expressed in neurons and activated by Ca²⁺-calmodulin following NMDAr activation [17].
- Endothelial NOS (eNOS): Constitutively expressed in endothelial cells, activated by shear stress or Ca²⁺-calmodulin, and critically involved in vascular homeostasis [17] [20].
- Inducible NOS (iNOS): Expressed in glial cells (e.g., astrocytes, microglia) primarily under inflammatory conditions; its activity is Ca²⁺-independent and produces •NO for prolonged periods [17].
- The Nitrate-Nitrite-NO Pathway: This NOS-independent pathway provides an important backup system for •NO generation, particularly under hypoxic conditions when NOS activity is limited by low O₂. Inorganic nitrate (NO₃⁻) from the diet is reduced to nitrite (NO₂⁻) by commensal bacteria or mammalian enzymes, which is then further reduced to •NO by various heme-containing proteins [17].
- Signaling and Bioavailability: The primary vasodilatory mechanism of •NO involves the activation of soluble guanylate cyclase (sGC) in vascular smooth muscle cells, leading to increased cyclic guanosine monophosphate (cGMP) and subsequent vasodilation [17] [19]. The bioavailability of •NO is critically threatened by oxidative stress, as •NO rapidly reacts with superoxide anion (O₂•⁻) to form peroxynitrite (ONOO⁻), a cytotoxic molecule that can uncouple NOS enzymes, further exacerbating oxidative damage [17].
Prostaglandins and Other Eicosanoids
Prostaglandins (PGs), vasoactive lipids derived from arachidonic acid, constitute another major signaling pathway in NVC. Their synthesis is initiated when neural activity triggers an increase in intracellular Ca²⁺ in astrocytes, activating the enzyme phospholipase A₂ (PLA₂). PLA₂ releases arachidonic acid from membrane phospholipids, which is then metabolized by cyclooxygenase (COX), particularly the COX-2 isoform, to produce prostaglandin H₂ (PGH₂), the precursor for various prostanoids, including the vasodilator PGE₂ [1] [11]. A recent multidisciplinary study identified PGE₂ as the principal prostaglandin in NVC, with pyramidal neurons being a major cellular source, and its vasodilatory effects are mediated primarily through the EP2 and EP4 receptors on vascular smooth muscle cells [11]. It is important to note that astrocytes can also produce vasoconstrictive arachidonic acid metabolites, suggesting a complex, finely-tuned regulation of vascular tone [1].
Potassium Ions
The regulated efflux of potassium ions (K⁺) from neurons and astrocytes is a key mechanism for mediating vasodilation. During neural activity, a local increase in extracellular K⁺ concentration can hyperpolarize vascular smooth muscle cells by activating inward-rectifier potassium (KIR) channels and sodium-potassium ATPase (Na⁺/K⁺-ATPase) pumps [1]. This hyperpolarization leads to the closure of voltage-gated calcium channels, reducing intracellular Ca²⁺ and promoting vasodilation. Astrocytes contribute significantly to this process; their endfeet, which enwrap cerebral blood vessels, are enriched with BKCa (large-conductance Ca²⁺-activated K⁺) channels. Astrocytic Ca²⁺ elevations can activate these channels, resulting in a targeted release of K⁺ into the perivascular space to modulate arteriolar diameter [1].
Integrated Signaling Pathways
The vasoactive signals described above do not operate in isolation but form an integrated, hierarchical network to ensure a robust and spatially precise hemodynamic response. The following diagram illustrates the primary cellular pathways and their interactions.
Figure 1. Integrated cellular pathways of vasoactive signaling in neurovascular coupling. Glutamate release from neurons acts on both neuronal NMDA receptors and astrocytic metabotropic glutamate receptors (mGluRs), initiating parallel signaling cascades that converge to cause vasodilation. Key: NO (Nitric Oxide), PGE₂ (Prostaglandin E₂), K⁺ (Potassium Ions).
The process begins with glutamatergic synaptic activity [17]. The canonical pathway involves the activation of neuronal NMDAr, Ca²⁺ influx, and nNOS-derived NO production, which is considered a major contributor to the rapid vasodilation [11]. In parallel, glutamate activation of astrocytic mGluRs elevates intracellular Ca²⁺ in astrocytes, leading to the production of PGE2 and the efflux of K⁺ from their endfeet [1] [11]. While the neuronal pathway is associated with the large and rapid component of vasodilation, the astrocytic pathway may be involved in slower, more sustained regulation of blood flow [1]. These signals collectively act on vascular smooth muscle cells to induce hyperpolarization and relaxation, thereby increasing local blood flow to match the metabolic demand of the active neurons.
Quantitative Data and Experimental Evidence
Table 2: Quantitative Effects of Vasoactive Signaling Modulation on Hemodynamic Responses
| Intervention / Condition | Experimental Model | Key Measured Outcome | Reported Effect | Citation |
|---|---|---|---|---|
| nNOS Inhibition | In vivo (Multiple animal studies) | Neurovascular Response (CBF) | Average reduction of 64% (across 11 studies) | [11] |
| NOS/COX/Epoxygenase Inhibition | Young C57BL/6 mice | Gait Coordination (Duty Cycle) | Significant decrease; altered footfall patterns | [21] |
| History of mTBI | Retired rugby players (Human, fNIRS) | Oxyhemoglobin (O₂Hb) in Left MFG | Reduced response: -0.015 ± 0.258 μM vs -0.160 ± 0.311 μM in controls | [8] |
| Cognitive-Motor Dual-Task | Healthy humans (EEG-fNIRS) | Neurovascular Coupling (NVC) Strength | Decreased NVC in theta, alpha, and beta EEG rhythms | [18] |
| Functional-Pharmacological Coupling (MPH) | Humans with ADHD | Sustained Attention Performance | Improved performance when MPH coupled with cognitive task | [22] |
Research Reagent Solutions and Methodologies
A rigorous investigation of vasoactive signaling requires a toolkit of specific pharmacological agents, genetic models, and advanced imaging techniques.
Table 3: Essential Research Reagents and Methodologies for Investigating Vasoactive Signaling
| Category / Target | Example Reagents / Tools | Primary Function / Mechanism | Application Note |
|---|---|---|---|
| NO Signaling | L-NAME (NOS inhibitor) | Non-selective inhibitor of NOS enzymes; reduces •NO production. | Used to establish causal role of NO; mimics NVC aspects of aging [21]. |
| 7-NI (7-Nitroindazole) | Relatively selective inhibitor of nNOS. | Used to dissect neuronal vs. endothelial NO contributions [17]. | |
| Prostaglandin Signaling | Indomethacin | Non-selective cyclooxygenase (COX) inhibitor. | Blocks prostaglandin synthesis; used to assess their contribution to NVC [21]. |
| Glutamate Receptors | MK-801 | Non-competitive NMDAr antagonist. | Blocks initial trigger for neuronal NO pathway [17]. |
| Potassium Channels | BaCl₂ (Barium Chloride) | Inhibitor of inward-rectifier K⁺ (KIR) channels. | Used to investigate K⁺-mediated vasodilation [1]. |
| Experimental Models | Genetic Knockout Mice (e.g., nNOS⁻/⁻, eNOS⁻/⁻) | Models with specific gene deletions. | Allows for dissection of specific isoform functions in NVC [17] [11]. |
| Imaging & Measurement | Laser Speckle Contrast Imaging | Measures cortical blood flow changes in vivo. | High spatial and temporal resolution for monitoring CBF [1]. |
| fNIRS / fMRI / EEG | Non-invasive human neuroimaging. | Assesses integrated NVC response and its impairment in pathology [8] [18]. |
Detailed Experimental Protocol: Pharmacological Dissection of NVC
The following workflow, derived from classic and contemporary studies, outlines a standard approach for isolating the contribution of different vasoactive pathways in an animal model [21].
Figure 2. Experimental workflow for pharmacological dissection of NVC pathways. This protocol allows for the quantitative assessment of the contribution of specific vasoactive pathways (NO, prostaglandins, EETs) to the overall functional hyperemia response.
Procedure:
- Baseline Measurement: In an anesthetized or awake, head-fixed animal model (e.g., mouse), a functional hyperemia response is elicited using a controlled stimulus (e.g., whisker stimulation, forepaw stimulation, or visual stimulus). The resulting change in local CBF is measured using a high-resolution technique such as laser Doppler flowmetry or laser speckle contrast imaging [1] [21].
- Pharmacological Inhibition: Animals are systemically (e.g., intraperitoneal injection) or locally (e.g., topically on the cortex) administered with one or more pathway-specific inhibitors.
- Post-Inhibition Measurement: After a suitable period for drug action, the exact same functional stimulus is applied, and the CBF response is measured again.
- Data Analysis: The post-inhibition CBF response is quantitatively compared to the baseline response. The percentage reduction in the response amplitude (e.g., peak CBF change) directly indicates the relative contribution of the inhibited pathway to the total NVC response. This method has shown that nNOS inhibition can reduce the response by an average of 64% [11].
Implications for Functional Neuroimaging and Disease
The vasoactive signaling pathways detailed above form the biological foundation for non-invasive neuroimaging techniques like fMRI and fNIRS [1] [18]. The blood-oxygen-level-dependent (BOLD) signal in fMRI is an indirect measure of neural activity that is heavily influenced by the neurovascular response and the resulting changes in blood flow, blood volume, and oxygen metabolism [17] [1]. Therefore, any pathology or pharmacological manipulation that alters the efficacy of glutamate, NO, prostaglandin, or potassium signaling can directly impact the BOLD signal, potentially confounding its interpretation as a pure index of neural activity [17] [21] [22].
Dysregulation of NVC is a hallmark of numerous neurological conditions. In Alzheimer's disease, amyloid-β peptides can disrupt NVC, potentially by inducing oxidative stress that scavenges NO and promotes peroxynitrite formation, leading to endothelial and neuronal dysfunction [17] [1]. In cerebral small vessel disease and hypertension, chronic oxidative stress and inflammation impair NO bioavailability, leading to neurovascular uncoupling [1] [11]. Even mild traumatic brain injury (mTBI), as seen in retired athletes, can lead to long-term impairments in hemodynamic responses, as evidenced by blunted O₂Hb measured with fNIRS [8]. Furthermore, experimental induction of neurovascular uncoupling in mice leads to measurable deficits in complex motor behaviors like gait coordination, establishing a direct cause-and-effect relationship between NVC failure and functional impairment [21].
Emerging therapeutic strategies focus on rescuing NO bioavailability through dietary interventions (e.g., nitrate and polyphenols) or physical exercise, which may help mitigate NVC dysfunction in neuropathological conditions [17]. Moreover, the novel concept of "functional-pharmacological coupling" proposes that administering a drug alongside a behavioral task that activates the drug's target brain circuits can enhance drug delivery and efficacy by leveraging activity-dependent increases in local cerebral blood flow [22]. This approach highlights the potential for harnessing our understanding of NVC for improved pharmacotherapy.
The Hemodynamic Response Function (HRF) is a fundamental physiological concept that describes the tightly regulated temporal relationship between local neural activity and subsequent changes in cerebral blood flow. This relationship forms the cornerstone of non-invasive brain imaging techniques, most notably functional Magnetic Resonance Imaging (fMRI) and functional Near-Infrared Spectroscopy (fNIRS), which rely on blood flow changes as a proxy for neural activity [23] [24]. In healthy adults, an increase in neuronal firing triggers a complex cascade of events leading to a localized influx of oxygenated blood, a process known as functional hyperemia [25]. This response is not merely a metabolic support function; emerging evidence suggests it may play an active role in modulating neural circuitry and information processing [25]. Understanding the precise shape, timing, and determinants of the HRF is therefore critical for accurate interpretation of neuroimaging data across basic research and clinical applications, including drug development [26].
This guide provides an in-depth technical examination of the HRF, framing it within the broader context of neurovascular coupling research. It details the biological mechanisms, methodological considerations for measurement, and critical sources of variability that researchers and drug development professionals must account for in their experimental design and data analysis.
Core Neurovascular Coupling Mechanisms
The HRF is the observable output of neurovascular coupling (NVC), the biological process orchestrated by the neurovascular unit (NVU). The NVU is an integrated entity comprising neurons, astrocytes, vascular smooth muscle cells (VSMCs), and pericytes [13] [27]. Signaling within this unit ensures that active brain regions receive a rapid and precise supply of energy substrates.
Molecular Signaling Pathways
The communication between neurons, glia, and vasculature involves several overlapping and redundant molecular pathways, which are summarized in the diagram below.
- Nitric Oxide (NO) Pathway: Neuronal depolarization leads to calcium influx, activating neuronal nitric oxide synthase (nNOS). The produced NO is a potent vasodilator. It exerts its effects on vascular smooth muscle cells by raising cyclic guanosine monophosphate (cGMP) levels. Evidence suggests that in capillary pericytes, NO may act indirectly by inhibiting the production of the vasoconstrictor 20-HETE [13].
- Arachidonic Acid Metabolites: Glutamate-mediated activation of astrocytic metabotropic receptors triggers calcium increases, leading to the production of arachidonic acid (AA). AA is metabolized into vasoactive agents, including prostaglandin E2 (PGE2) and epoxyeicosatrienoic acids (EETs), which act on the prostaglandin E2 receptor 4 (EP4) on pericytes and VSMCs to induce vasodilation [13] [23].
- Purinergic and Other Signals: Neuronal activity also leads to the release of adenosine, which acts as a vasodilator, particularly when neuronal ATP is low [23]. Potassium ions and other vasoactive molecules also contribute to this complex, redundant signaling system [13].
The result of these pathways is the relaxation of VSMCs and capillary pericytes, leading to vasodilation and a pronounced increase in local cerebral blood flow that substantially exceeds the immediate metabolic oxygen demand [23]. This oversupply is the physiological basis for the Blood Oxygen Level Dependent (BOLD) contrast used in fMRI [25] [23].
Quantitative Profiling of the HRF
The canonical HRF is a stereotypical waveform characterized by several key parameters that can be quantified. These metrics are essential for modeling and interpreting BOLD and fNIRS signals.
Table 1: Key Quantitative Parameters of the Hemodynamic Response Function
| Parameter | Typical Value (fMRI/BOLD) | Description | Physiological Significance |
|---|---|---|---|
| Onset (Latency) | 1-2 seconds | Delay between neural impulse and the start of the HRF. | Speed of neurovascular signaling. |
| Time-to-Peak (TTP) | 5-6 seconds | Time taken for the HRF to reach its maximum amplitude. | Efficacy of the vasodilatory response. |
| Full-Width at Half-Maximum (FWHM) | 4-5 seconds | Width of the HRF at half of its peak amplitude. | Duration of the hemodynamic response. |
| Response Height (RH) | 0.5-2% BOLD change | Peak amplitude of the HRF. | Strength of the blood flow response. |
| Undershoot | ~10% of peak | A negative dip following the main response. | Post-stimulus vasoconstriction or metabolic processes. |
These parameters are not fixed. A 2025 study analyzing high-resolution fMRI data from the Human Connectome Project found significant variability in HRF amplitude and latency across different brain regions and tasks, while TTP and FWHM were relatively more consistent [28]. This variability must be modeled for accurate analysis.
Mathematical Modeling of the HRF
In practice, the HRF is often modeled mathematically to serve as a regressor in statistical models like the General Linear Model (GLM). The most common model uses a combination of two Gamma functions to capture the positive response and the subsequent undershoot [29]:
HRF(t) = A * ( (t^(α₁-1) * β₁^α₁ * exp(-β₁*t)) / Γ(α₁) - c * (t^(α₂-1) * β₂^α₂ * exp(-β₂*t)) / Γ(α₂) )
Where:
Ais a scaling factor for amplitude.α₁, β₁control the delay and dispersion of the positive response.α₂, β₂control the delay and dispersion of the undershoot.cis a scaling factor for the undershoot ratio.Γis the Gamma function.
This model typically involves six free parameters that can be estimated from data to account for inter-regional and inter-subject variability [29]. More flexible approaches, such as using Fourier basis sets or sine series expansions, are also employed to capture even greater shape variability without assuming a rigid canonical form [28].
Methodological Approaches for HRF Investigation
Different neuroimaging modalities provide unique windows into the HRF, each with advantages and limitations.
Primary Imaging Modalities
Table 2: Key Modalities for Measuring the Hemodynamic Response
| Modality | Measured Signal | Spatial Resolution | Temporal Resolution | Key Advantages & Applications |
|---|---|---|---|---|
| fMRI | Blood Oxygenation Level Dependent (BOLD) | High (mm) | Moderate (1-2 s) | Gold standard for whole-brain mapping; excellent spatial resolution [26]. |
| fNIRS | Concentration changes in Oxy-Hb and Deoxy-Hb | Low (cm) | High (~0.1 s) | Portable, tolerant of movement; ideal for naturalistic settings, bedside monitoring, and BCI [29] [30]. |
| Arterial Spin Labeling (ASL) fMRI | Cerebral Blood Flow (CBF) | High (mm) | Low (several s) | Provides quantitative CBF measurement, not just a relative signal like BOLD [26]. |
Experimental Protocols for HRF Assessment
Robust experimentation requires carefully designed protocols. Below is a generalized workflow for an HRF study, integrating elements from both fMRI and fNIRS approaches.
Detailed Methodological Notes:
- Stimulus Paradigms: For precise HRF estimation, event-related designs with brief, isolated stimuli are often preferred over block designs. The duration and intensity of the stimulus should be carefully controlled [28].
- fNIRS Pre-processing: Raw fNIRS signals require specific pre-processing to remove physiological noise. This often involves wavelet transform techniques to isolate the task-evoked signal from cardiac pulsation, respiratory signals, and low-frequency Mayer waves [29] [31].
- HRF Estimation in GLM: The General Linear Model framework is used to estimate the HRF's contribution to the measured signal. The model is formulated as:
Y = X * β + εwhereYis the measured BOLD or fNIRS signal,Xis the design matrix containing the convolved HRF model,βis the vector of unknown weights (estimating activation strength), andεis the error term [29] [31]. Model fit can be improved by adding time and dispersion derivatives to the canonical HRF to account for slight timing and shape variations [24]. - Deconvolution Methods: When the precise timing of neural events is unknown (e.g., in resting-state fMRI), blind deconvolution techniques can be employed. These methods, such as the data-driven
rsHRFtoolbox or stochastic Dynamic Causal Modeling (DCM), solve the inverse problem to estimate both the neural activity and the HRF shape directly from the fMRI time series [24].
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 3: Key Reagents and Tools for HRF Research
| Item / Reagent | Function / Role | Example Use Case |
|---|---|---|
| Canonical HRF Model | A fixed, standardized model of the hemodynamic response. | Serves as the default regressor in GLM for task-based fMRI/fNIRS analysis [29]. |
| General Linear Model (GLM) | A statistical framework for estimating the contribution of the HRF to the measured signal. | Used to compute statistical parametric maps of brain activation [29] [31]. |
| NIRS-SPM Toolbox | A public statistical toolbox for fNIRS data analysis. | Provides a validated pipeline for fNIRS data preprocessing and GLM-based activation mapping [29]. |
| Finite Impulse Response (FIR) Basis Set | A flexible set of basis functions that does not assume a fixed HRF shape. | Used to estimate the shape of the HRF in a data-driven manner with minimal assumptions [28]. |
| Pharmacological Agents (e.g., NOS inhibitors) | Used to probe specific neurovascular coupling pathways. | Administered in animal models to isolate the contribution of nitric oxide to the HRF [13]. |
| Wavelet Transform Toolboxes | Algorithms for decomposing signals into time-frequency space. | Applied to fNIRS data to isolate and remove physiological noise (cardiac, respiratory) [29] [31]. |
| Simplex/Nelder-Mead Algorithm | A nonlinear optimization algorithm for parameter estimation. | Used to find the optimal parameters (e.g., delays, dispersions) for a flexible HRF model in fNIRS [29]. |
Critical Considerations and Clinical Relevance
Acknowledging and accounting for HRF variability is paramount for robust research and clinical application.
The HRF is not a one-size-fits-all function. Its shape varies significantly due to:
- Regional Differences: HRF shape differs across brain regions, likely due to local variations in vascular density and neurovascular architecture [28] [24]. For example, the HRF in the visual cortex has been shown to be parametrically different from that in the motor cortex [28].
- Inter-Subject Variability: Substantial differences in HRF shape exist between individuals, sometimes exceeding regional differences within a single subject [29] [24]. Time-to-peak can vary by up to several seconds across individuals [28].
- Developmental and Aging Factors: The HRF matures postnatally. In infants, neural activation can evoke a negative BOLD response, indicating an immature NVU where blood flow does not properly match metabolic demand. This response invertes to the canonical positive shape as astrocytes and the vascular network mature [27]. Aging can also alter HRF properties [28].
- Pathological Alterations: Diseases that affect the neurovascular unit profoundly disrupt the HRF. In Alzheimer's Disease, neurovascular uncoupling is a prominent early feature, driven by amyloid-β pathology and astrocyte dysfunction [13] [27]. Similar disruptions are observed in stroke and other neurodegenerative conditions [27].
- Physiological Confounds: Non-neural factors like blood composition (hematocrit), caffeine/alcohol intake, and cardiorespiratory cycles can modulate the HRF shape and confound neuroimaging results [24].
Ignoring HRFv, particularly in resting-state functional connectivity studies, can lead to severe confounds. Apparent correlations or group differences in connectivity can be driven by vascular differences rather than true neural synchrony [24]. For instance, one study noted that HRF differences between women and men led to a 15.4% median error in functional connectivity estimates in a group-level comparison [24].
Application in Drug Development
fMRI and fNIRS have growing roles in the drug development pipeline, where the HRF serves as a critical link between molecular intervention and systems-level brain function.
- Target Engagement and Pharmacodynamics: An fMRI signal can provide indirect evidence of target engagement if a biologically plausible link exists between the drug's molecular target and the resulting hemodynamic change [26]. For example, a drug targeting the NO signaling pathway should manifest as an altered HRF in response to a cognitive task.
- Dose-Response Relationships: fMRI can be used in Phase I trials to establish dose-response and exposure-response relationships, guiding dose selection for later-phase trials [26].
- Biomarker Qualification: While no fMRI biomarker has yet been fully qualified by regulatory agencies like the FDA or EMA, consortia are actively seeking qualification for specific contexts of use, such as stratifying patients with autism spectrum disorder [26].
The Hemodynamic Response Function is more than a mere link between neural activity and blood flow; it is a dynamic and variable reflection of a complex biological process. A deep understanding of its underlying mechanisms, sources of variability, and appropriate measurement methodologies is essential for any researcher or professional using fMRI or fNIRS. As the field moves towards precision mental health and individualized biomarkers, the ability to account for individual HRF signatures—through dense sampling [30], advanced deconvolution [24], and personalized modeling [28]—will be crucial for developing accurate diagnostics and effective, personalized therapeutics for neurological and psychiatric disorders.
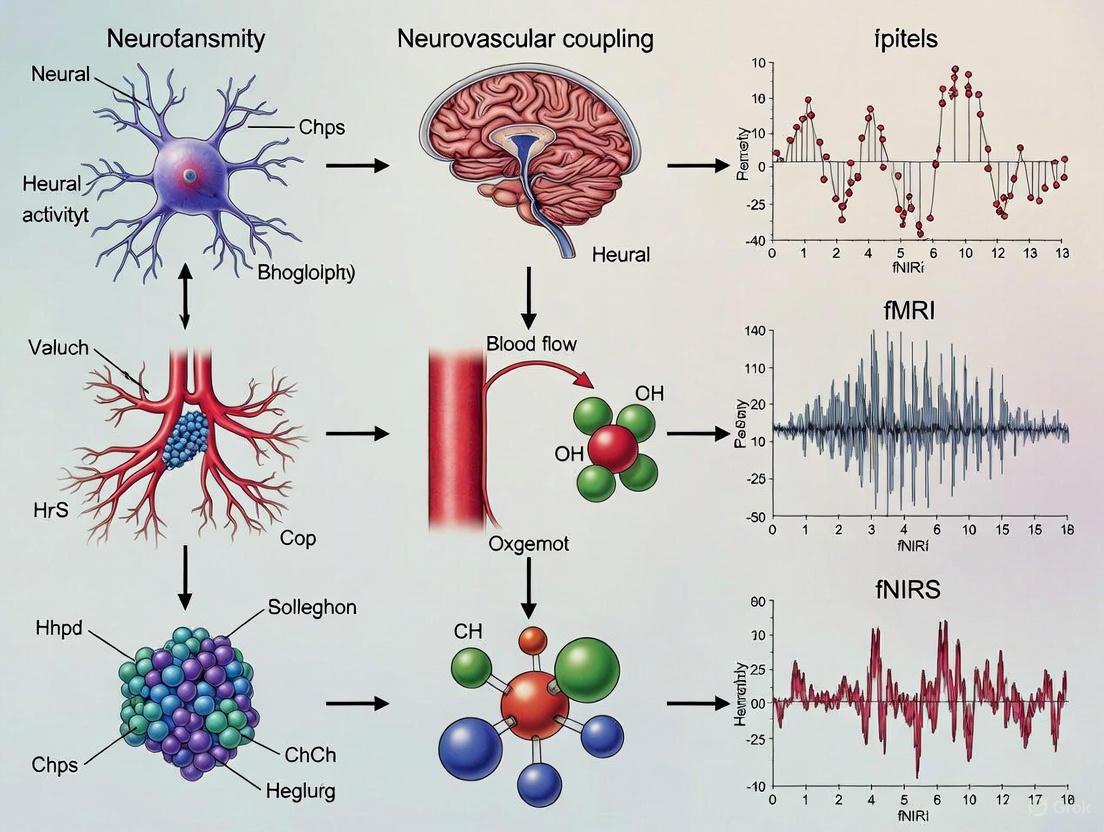
Neurovascular Coupling as the Foundation for fMRI and fNIRS Signals
Neurovascular coupling (NVC) describes the fundamental physiological process whereby neuronal activity triggers localized changes in cerebral blood flow (CBF), a mechanism critical for interpreting functional neuroimaging signals [2]. First articulated by Roy and Sherrington in 1890 as the "intrinsic regulation of local CBF," this relationship ensures that active brain regions receive adequate oxygen and nutrients to meet metabolic demands [2]. The neurovascular unit (NVU)—an integrated system comprising neurons, astrocytes, vascular smooth muscle cells, and pericytes—orchestrates this precise coordination [13] [2]. Functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS), two cornerstone non-invasive neuroimaging technologies, rely entirely on measuring hemodynamic changes consequent to NVC. The fMRI signal primarily reflects the Blood Oxygenation Level-Dependent (BOLD) contrast, arising from local changes in deoxyhemoglobin concentration [1] [2]. fNIRS directly measures relative concentration changes of oxyhemoglobin (O2Hb) and deoxyhemoglobin (HHb) in the cortical microvasculature [32] [33]. Understanding NVC is therefore not merely academic but essential for accurately interpreting the neural significance of data acquired with these widespread modalities.
Molecular Mechanisms of Neurovascular Coupling
The signaling pathways of NVC involve a complex, interacting cascade of vasoactive molecules released by neurons and astrocytes in response to synaptic activity.
Key Signaling Pathways and Vasoactive Agents
Upon neuronal activation, neurotransmitters like glutamate are released, initiating a multi-pathway response. Nitric oxide (NO), a potent vasodilator, is synthesized in neurons by neuronal nitric oxide synthase (nNOS) following calcium influx. NO raises cyclic guanosine monophosphate (cGMP) levels in vascular smooth muscle cells (VSMCs), leading to relaxation and vasodilation [13]. Arachidonic acid metabolites constitute another major pathway. Phospholipase D2 (PLD2) initiates arachidonic acid synthesis, which is subsequently metabolized by cyclooxygenase-1 (COX1) into the vasodilatory prostaglandin E2 (PGE2). PGE2 acts on the EP4 receptor, a Gs-linked G-protein-coupled receptor on capillary pericytes and VSMCs, increasing intracellular cAMP and causing vasodilation [13]. Potassium ions (K+) also play a crucial role; they are released into the perivascular space through large-conductance calcium-activated potassium (BK) channels on astrocytic endfeet, which envelop arterioles. Moderate perivascular K+ concentration (5–20 mM) induces hyperpolarization and dilation of VSMCs [2]. Furthermore, astrocytic calcium signaling is pivotal. Increases in astrocytic Ca2+ can trigger the release of vasoactive agents, with moderate increases promoting dilation and larger spikes potentially causing constriction [2]. These pathways exhibit regional heterogeneity and work in concert, as inhibiting any single pathway does not completely abolish the hemodynamic response [13].
Figure 1: Key Cellular Signaling Pathways in Neurovascular Coupling. The diagram illustrates the major vasodilatory pathways initiated by neuronal activity, involving neurons and astrocytes acting on vascular smooth muscle cells and pericytes. NO: Nitric Oxide; nNOS: neuronal Nitric Oxide Synthase; sGC: soluble Guanylate Cyclase; cGMP: cyclic Guanosine Monophosphate; BK: Large-conductance Calcium-activated Potassium channel; PGE2: Prostaglandin E2; EETs: Epoxyeicosatrienoic Acids; AA: Arachidonic Acid; COX1: Cyclooxygenase-1.
The Hemodynamic Response Function
The net effect of these molecular pathways is the hemodynamic response function (HRF), the characteristic temporal pattern of blood flow and oxygenation changes measured by fMRI and fNIRS. Following neuronal activation, a tightly coordinated sequence occurs: a localized increase in cerebral blood flow manifests after approximately a 2-second delay, peaks around 4-6 seconds post-stimulus, and is followed by a slow return to baseline, sometimes accompanied by a post-stimulus undershoot [32] [2]. This hemodynamic response is characterized by a local increase in O2Hb and a decrease in HHb due to an overcompensatory delivery of oxygenated blood [32] [8]. The BOLD signal in fMRI is sensitive to the resulting decrease in the concentration of paramagnetic deoxyhemoglobin [13] [2]. It is crucial to recognize that the HRF is an indirect and delayed measure of neural activity, reflecting a complex integration of synaptic input and local processing more closely than direct neuronal spiking output [2].
Experimental Evidence and Quantitative Data
Empirical studies robustly demonstrate the intensity-dependent nature of neurovascular responses and the validity of combining electrophysiological and hemodynamic measurements.
Intensity-Dependent Amplitude Changes
Research using integrated EEG-fNIRS paradigms has quantitatively linked electrophysiological activity to hemodynamic changes. One study presenting auditory tones of varying intensities (70.9 dB to 94.5 dB) found that increases in tone intensity led to graded enhancements in EEG event-related potential (ERP) components (N1, P2, and N1-P2 peak-to-peak amplitude) [32]. Concurrently, fNIRS measurements showed amplitude increases in O2Hb and decreases in HHb in the auditory and prefrontal cortices [32]. Spearman correlation analysis specifically identified a relationship between the left auditory cortex and N1 amplitude, and the right dorsolateral cortex with P2 amplitude, particularly for HHb concentrations. These findings provide direct evidence for neurovascular coupling by demonstrating a systematic relationship between the amplitude of electrical and hemodynamic responses to sensory stimulation [32].
Table 1: Summary of Intensity-Dependent Neurovascular Responses to Auditory Stimulation
| Experimental Parameter | EEG/ERP Findings | fNIRS Findings | Correlation Evidence |
|---|---|---|---|
| Stimulus Intensity | Increased N1, P2, and N1-P2 peak-to-peak amplitude [32] | Increased O2Hb; Decreased HHb in auditory & prefrontal cortices [32] | Significant Spearman correlations between ERP amplitudes (N1, P2) and hemodynamic concentrations [32] |
| Stimulus Paradigm | Three-tone (77.9, 84.5, 89.5 dB) and five-tone intensities (70.9-94.5 dB) [32] | Hemodynamic changes observed in auditory and prefrontal cortices [32] | Left auditory cortex with N1 amplitude; Right dorsolateral cortex with P2 amplitude [32] |
fNIRS in Cognitive and Clinical Paradigms
fNIRS studies of cognitive-motor interference (CMI) further illuminate NVC in complex tasks. A study involving single and dual cognitive-motor tasks found that the extra cognitive load in the dual-task condition led to decreased neurovascular coupling between fNIRS and EEG signals across theta, alpha, and beta rhythms [18]. This suggests that divided attention can impair the efficiency of the neurovascular response. Clinically, NVC is often impaired; a study of retired rugby players with a history of concussion demonstrated a blunted hemodynamic response during a "Where's Wally" visual search task compared to controls. The control group showed a greater relative increase in O2Hb, while the mTBI group exhibited a reduced O2Hb response and a greater rate of oxygen extraction, indicating altered cerebral metabolic demands following injury [8].
Table 2: Neurovascular Coupling Findings in Cognitive and Clinical Studies
| Study Paradigm | Population | Key NVC Findings |
|---|---|---|
| Cognitive-Motor Interference (CMI) [18] | 16 healthy young adults | Decreased EEG-fNIRS coupling in dual-task vs. single-task in theta, alpha, and beta rhythms [18] |
| Sport-Related Concussion (mTBI) [8] | 21 retired rugby players vs. 23 controls | Reduced O2Hb increase in left middle frontal gyrus in mTBI group; Indicated altered metabolic demand [8] |
| Sleep Inertia [34] | 21 healthy adults | Dynamic, time-varying coupling between EEG alpha/vigilance and fMRI BOLD in thalamus/ACC post-awakening [34] |
Methodologies for Investigating Neurovascular Coupling
Experimental Protocols and Designs
Investigating NVC requires carefully designed protocols that can elicit and measure coupled neural and vascular activity.
Auditory Intensity Paradigm: A classic protocol for testing NVC involves presenting participants with tones of different intensities. One experiment used three-tone intensities (77.9 dB, 84.5 dB, and 89.5 dB), each lasting 500 ms and randomly presented 54 times. A second experiment used five intensities (70.9 dB to 94.5 dB) presented in trains of 8 tones. Throughout stimulation, EEG records ERPs (N1, P2), while fNIRS measures hemodynamic changes in the auditory, visual, and prefrontal cortices. This design directly tests the amplitude dependence of both signals and their correlation [32].
Cognitive-Motor Dual-Task Paradigm: To study NVC under cognitive load, experiments can be designed with single motor, single cognitive, and cognitive-motor dual tasks. For example, a grip force tracking task (motor) and a number detection task (cognitive) can be performed separately and concurrently. During these tasks, EEG and fNIRS are recorded simultaneously. The extracted task-related components from both modalities are then analyzed for their correlation to quantify NVC strength, which typically decreases under dual-task conditions [18].
The "Where's Wally" NVC Test: This visual cognitive task is a validated measure of NVC, useful for clinical populations. Participants sit quietly for a 5-minute baseline, then complete five cycles of a task. Each cycle consists of 20 seconds with eyes closed followed by 40 seconds of visually searching for the character "Wally" in a complex image. The fNIRS system, placed over the prefrontal cortex, monitors changes in O2Hb and HHb throughout. This protocol effectively evokes a measurable hemodynamic response related to visual search and attention [8].
Figure 2: Generalized Workflow for Neurovascular Coupling Experiments. The flowchart outlines the common steps in a NVC investigation, from baseline recording and stimulus presentation to concurrent multi-modal signal acquisition and final analysis.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Reagents and Materials for NVC Research
| Item/Tool | Primary Function in NVC Research |
|---|---|
| fNIRS System (e.g., multi-channel systems like OctaMon) | Non-invasive, silent measurement of relative O2Hb and HHb concentration changes in the cerebral cortex [32] [8]. |
| EEG System (MR-compatible for simultaneous fMRI) | High-temporal-resolution recording of electrical brain activity (ERPs, spectral power) for correlation with hemodynamics [32] [34]. |
| fMRI Scanner | Mapping brain activation via the BOLD signal, an indirect measure of neural activity based on NVC [1] [2]. |
| Pharmacological Agents (e.g., NOS inhibitors, COX inhibitors) | Selective blockade of specific vasoactive pathways (e.g., NO, PGE2) to dissect their contribution to the hemodynamic response [13]. |
| Task-Related Software | Presentation of controlled sensory, motor, or cognitive stimuli (e.g., auditory tones, "Where's Wally" task) to evoke calibrated neural activity [32] [8]. |
| Computational Models (e.g., Balloon model, Dynamic Causal Modeling) | Biophysical models that link neuronal activity to BOLD/fNIRS signals, allowing inference of neural processes from hemodynamic data [1] [35]. |
Neurovascular coupling is the indispensable biological link that allows researchers to infer brain function from hemodynamic signals measured by fMRI and fNIRS. The process is governed by a sophisticated interplay of cellular and molecular mechanisms within the neurovascular unit, resulting in a characteristic hemodynamic response. Quantitative evidence firmly establishes a correlation between the amplitude of electrophysiological events and subsequent vascular changes. Methodologies combining EEG with fNIRS or fMRI are powerful tools for probing this relationship in both healthy and diseased states. A deep understanding of NVC principles is therefore paramount for the correct design, analysis, and interpretation of functional neuroimaging studies across basic neuroscience and clinical drug development.
Methodological Approaches: Applying fMRI and fNIRS to Measure NVC in Basic and Clinical Research
The Blood-Oxygen-Level-Dependent (BOLD) signal, the primary contrast mechanism for functional magnetic resonance imaging (fMRI), provides an indirect window into brain function by detecting hemodynamic changes coupled to neural activity. This technical guide details the physiological origin of the BOLD signal within the framework of neurovascular coupling (NVC), the functional hyperemia that links neuronal firing to localized blood flow and oxygenation changes. We summarize key biophysical parameters, present standardized experimental methodologies for its investigation, and visualize core signaling pathways. The document also places BOLD fMRI in the context of multimodal brain research, particularly in conjunction with functional near-infrared spectroscopy (fNIRS), and provides a practical toolkit of research reagents and solutions. This resource is intended for researchers, scientists, and drug development professionals seeking a current and in-depth understanding of BOLD fMRI fundamentals and applications.
Functional magnetic resonance imaging (fMRI) has revolutionized cognitive neuroscience and clinical brain mapping since its inception in the early 1990s [36]. The vast majority of fMRI studies rely on the Blood-Oxygen-Level-Dependent (BOLD) contrast, a non-invasive measure that allows for the visualization of brain activity by inferring regional neural activity from associated changes in blood oxygenation [37] [38]. The BOLD signal is fundamentally an indirect metric, arising from a complex physiological process known as neurovascular coupling (NVC). NVC describes the mechanism by which neural activity triggers a cascade of events leading to a precisely regulated increase in local cerebral blood flow (CBF), which in turn alters the concentration of oxygenated and deoxygenated hemoglobin [7] [39].
Understanding the precise origin and nature of the BOLD signal is critical for its accurate interpretation. While traditionally viewed as a correlate of local excitatory neural activity, recent evidence challenges and refines this perspective, suggesting a more dominant role for specific cell types and revealing that signal decreases are tightly linked to active suppression of spiking activity [37] [40] [38]. Furthermore, the BOLD signal is not a pure measure of neural activity but is influenced by a multitude of physiological factors, including age, cardiorespiratory function, and the integrity of the vascular system [41].
This guide provides an in-depth examination of the BOLD signal. It begins by elucidating its physiological basis, summarizes key quantitative parameters in structured tables, details experimental protocols for its study, and presents visualizations of core pathways. Finally, it explores the synergy between BOLD fMRI and other modalities like fNIRS within the broader context of neurovascular research.
Physiological Basis and Origin of the BOLD Signal
The BOLD signal is a complex, indirect reflection of neural activity, governed by the principles of neurovascular coupling. The following diagram outlines the primary pathway from neural activity to the measured fMRI signal.
Figure 1: The Neurovascular Coupling Pathway. This diagram illustrates the primary sequence of events from neural activity to the measurable BOLD fMRI signal. Key processes include neurovascular coupling, the hemodynamic response, and the resulting change in MR image contrast.
The Hemodynamic Response and BOLD Contrast
The process begins with local neural activity, encompassing both excitatory and inhibitory processes. This activity triggers neurovascular coupling, a process mediated by neurons, astrocytes, and vascular cells, leading to vasodilation and a marked increase in local cerebral blood flow (CBF) [7]. This increase in flow is typically greater than the local oxygen consumption, resulting in a net increase in oxygenated hemoglobin (HbO) and a relative decrease in deoxygenated hemoglobin (HbR) in the venous capillaries and draining venules [42].
Deoxyhemoglobin is paramagnetic and acts as an intrinsic contrast agent, distorting the local magnetic field and leading to a faster decay of the MR signal (reduced T2* relaxation time). The reduction in deoxyhemoglobin concentration during neural activity thus reduces this distortion, leading to a longer T2 and a stronger MR signal in T2-weighted images—the positive BOLD signal [42] [43].
Cellular Origins of the BOLD Signal
A longstanding assumption was that the BOLD signal primarily reflected input and processing from excitatory neurons. However, a paradigm-shifting model-driven meta-analysis suggests a more complex picture, concluding that inhibitory interneurons drive over 75% of the neurovascular response across studies, while the contribution from excitatory cells may be less than 20% [40]. This indicates that the BOLD signal is heavily weighted toward the activity of local inhibitory circuits.
Furthermore, the relationship between neuronal firing and BOLD signal direction is highly specific. A tight link has been demonstrated in the human association cortex: single neurons in a region showing BOLD activation selectively increase their spiking rate to a preferred stimulus (e.g., faces). Conversely, in an adjacent region showing BOLD deactivation to the same stimulus, the majority of face-selective neurons (over 95%) showed a significant decrease in spike rate, with about a third showing genuine suppression below baseline activity [37] [38]. This evidence strongly indicates that negative BOLD responses can be a direct correlate of active suppression of neural spiking.
Key Parameters and Quantitative Data
The BOLD signal and its underlying physiology can be characterized by several key parameters. The table below summarizes the core parameters of the BOLD response itself, while the subsequent table outlines key metrics related to cerebral metabolism that are often derived or used in conjunction with BOLD fMRI.
Table 1: Key Parameters of the BOLD fMRI Signal
| Parameter | Description | Typical Value / Range |
|---|---|---|
| Temporal Delay (Hemodynamic Response) | The lag between neural activity onset and the peak BOLD signal. | 4-6 seconds [36] |
| Signal Change | The magnitude of the BOLD signal increase during neural activity, expressed as percent change from baseline. | 1-5% at 3T |
| Spatial Specificity | The precision with which the BOLD signal localizes the neural activity; limited by the vascular architecture. | Millimeter level [36] |
| Temporal Resolution | The ability to resolve changes over time, limited by the slow hemodynamic response. | ~0.33-2 Hz sampling rate [36] |
| BOLD Signal Origin | The relative contribution of different cell types to the neurovascular response. | >75% from inhibitory interneurons; <20% from excitatory cells [40] |
| Negative BOLD | A signal decrease below baseline, often linked to a decrease in neural spiking activity. | Associated with ~96% of single neurons decreasing firing rate [38] |
Table 2: Key Cerebral Metabolic Parameters in BOLD and Related Studies
| Parameter | Description | Typical Baseline Values (Gray Matter) | Measurement Methods |
|---|---|---|---|
| Cerebral Blood Flow (CBF) | The volume of blood flow per unit brain mass per time. | ~50-60 mL/100g/min [42] | pCASL, Phase Contrast MRI |
| Oxygen Extraction Fraction (OEF) | The fraction of oxygen extracted from the blood by the brain tissue. | ~31% (e.g., 31 ± 5% in constrained qBOLD) [42] | qBOLD, TRUST, OxFlow, PET |
| Cerebral Metabolic Rate of O2 (CMRO2) | The rate of oxygen consumption by the brain. | ~150-160 μmol/100g/min | Calculated via Fick's Principle: CMRO2 = CBF · (SaO2 - SvO2) · Ca [42] |
| Venous Oxygen Saturation (SvO2) | The saturation of oxygen in venous blood. | ~55-65% (Derived from OEF) | T2-based oximetry (e.g., MOTIVE), Susceptometry-based oximetry (e.g., OxFlow) [42] |
Experimental Protocols and Methodologies
This section outlines key experimental approaches for investigating the BOLD signal and neurovascular coupling, ranging from direct neuronal correlation to advanced metabolic mapping.
Correlating BOLD fMRI with Single-Unit Recordings
Objective: To establish a direct relationship between BOLD signal changes (both activation and deactivation) and changes in the firing rate of single neurons in the human brain [37] [38].
Protocol:
- Participant Selection: Patients with clinical indications for intracerebral electrode implantation (e.g., refractory epilepsy) provide a unique opportunity for such studies.
- fMRI Acquisition: Prior to or following implantation, conduct BOLD fMRI using a block-design or frequency-tagging paradigm. For example, present images of faces and non-face objects in an alternating sequence to identify category-selective voxels.
- Electrophysiological Recording: Implant hybrid macro-microelectrodes in the target region (e.g., fusiform gyrus). Record single-unit activity from the same region while the patient views the same visual stimulus paradigm used in fMRI.
- Data Analysis:
- fMRI: Use Fast Fourier Transform (FFT) on the BOLD time course to identify voxels with significant power at the stimulus frequency. Calculate Z-scores to define areas of signal increase (positive Z) or decrease (negative Z).
- Single-Units: Apply FFT to the neural spiking data. Identify neurons with significant responses at the base stimulus frequency (general visual response) and the specific category-tagging frequency (category-selectivity). Categorize neurons as showing significant increases or decreases in firing rate to the preferred category.
Constrained Quantitative BOLD (qBOLD) for Metabolic Mapping
Objective: To perform calibration-free, voxel-wise mapping of the oxygen extraction fraction (OEF) and cerebral metabolic rate of oxygen (CMRO2) [42].
Protocol:
- MRI Data Acquisition: Acquire multi-parametric data required for the constrained qBOLD model:
- R2' Map: Derived from a multi-echo gradient-echo sequence to quantify the reversible transverse relaxation rate.
- Cerebral Blood Volume (CBV): Measured using a method like velocity-selective venous spin labeling (VS-VSL).
- Cerebral Blood Flow (CBF): Quantified via pseudo-continuous arterial spin labeling (pCASL).
- Quantitative Susceptibility Mapping (QSM): Acquired to account for non-heme iron contributions.
- Model Fitting: Implement the constrained qBOLD model, which uses the acquired R2' and CBV maps as prior constraints to decouple SvO2 and deoxygenated blood volume (DBV) in the QSM+qBOLD analysis, solving for voxel-wise OEF.
- CMRO2 Calculation: Apply Fick's principle (see Table 2) using the mapped OEF, measured CBF, and assumed arterial oxygen saturation (SaO2) to calculate CMRO2.
- Validation: Compare OEF and CMRO2 values against global oximetry methods like MOTIVE or OxFlow at baseline and in response to a physiological challenge (e.g., caffeine ingestion).
Simultaneous fMRI-fNIRS Integration
Objective: To leverage the high spatial resolution of fMRI with the superior temporal resolution and portability of fNIRS for a more comprehensive assessment of brain function [36].
Protocol:
- Hardware Setup: Use MRI-compatible fNIRS optodes. Place the optodes on the scalp over the cortical regions of interest prior to the participant entering the MRI scanner. Ensure proper shielding to minimize electromagnetic interference.
- Synchronous Data Acquisition: Simultaneously record BOLD fMRI and fNIRS signals (concentrations of HbO and HbR) during resting-state or task-based paradigms (e.g., motor or cognitive tasks).
- Data Processing and Fusion:
- fMRI Preprocessing: Conduct standard steps including realignment, normalization, and smoothing.
- fNIRS Preprocessing: Perform channel inspection, filtering of cardiac and respiratory noise, and conversion of optical density to HbO/HbR concentrations.
- Data Integration: Employ general linear models (GLM) to relate fNIRS hemodynamic responses to the BOLD signal. Alternatively, use multivariate pattern analysis or machine learning techniques to fuse the datasets and identify coupled spatiotemporal patterns of activation.
- Application: This integrated approach is particularly valuable for validating fNIRS signals, studying neurovascular coupling in naturalistic settings, and for clinical monitoring where portability is beneficial.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents and Materials for Neurovascular Coupling and BOLD fMRI Research
| Item | Function / Application | Example Use Case |
|---|---|---|
| Frequency-Tagging Visual Stimuli | To objectively and efficiently identify category-selective neural and BOLD responses. | Presenting images of faces and objects at different rhythmic frequencies (e.g., 6Hz vs. 1.2Hz) to identify face-selective regions [37] [38]. |
| Caffeine (as a Vasoactive Stimulus) | A vasoconstrictive agent used to provoke changes in cerebral blood flow and metabolism. | Administering a 200mg caffeine pill to study the brain's metabolic and vascular response, observing increased OEF and reduced CBF with stable CMRO2 [42]. |
| Heart Rate Variability Biofeedback (HRV-BF) | A non-invasive breathing training to modulate autonomic function and brain-body coupling. | Investigating the impact of HRV-BF on age-related declines in heart rate-brain signal coupling, showing a shift towards a more youthful pattern in older adults [41]. |
| Optogenetic Constructs | To selectively activate or inhibit specific neuronal cell types (e.g., inhibitory interneurons) in animal models. | Probing the specific contributions of different neuronal populations to the hemodynamic response and BOLD signal [40]. |
| Constrained qBOLD Analysis Pipeline | A software and algorithmic suite for processing multi-parametric MRI data to compute OEF and CMRO2. | Enabling non-invasive, voxel-wise mapping of cerebral oxygen metabolism for studies on neurological disorders [42]. |
| MRI-compatible fNIRS System | Hardware for simultaneous acquisition of hemodynamic signals from fNIRS and fMRI. | Validating fNIRS measurements, improving temporal resolution of fMRI studies, and enabling brain imaging in more naturalistic contexts [36]. |
The fMRI-BOLD signal remains a pivotal tool for non-invasive human brain mapping, but its interpretation requires a nuanced understanding of its physiological origin. The signal is an indirect and complex reflection of neural activity, predominantly shaped by inhibitory interneuron-driven neurovascular coupling and resulting hemodynamic changes. Quantitative techniques like constrained qBOLD are advancing our ability to extract metabolic parameters, while multimodal integration with fNIRS is enriching the spatiotemporal characterization of brain function. A solid grasp of these principles, parameters, and methodologies is essential for researchers and drug development professionals to design robust experiments, accurately interpret BOLD data, and advance our understanding of brain function in health and disease.
Functional Near-Infrared Spectroscopy (fNIRS) has emerged as a powerful, non-invasive neuroimaging technique that measures cortical hemodynamics by detecting changes in oxyhemoglobin (HbO) and deoxyhemoglobin (HbR) concentrations. This technology leverages the fundamental physiological process of neurovascular coupling (NVC), the intricate mechanism whereby neural activity triggers localized changes in cerebral blood flow [8] [32]. When a brain region becomes active, its metabolic demands increase, leading to a complex vascular response that delivers oxygenated blood. fNIRS captures this response by measuring the characteristic hemodynamic changes associated with brain activation, providing a critical window into brain function [44].
The significance of fNIRS is particularly evident when framed within broader neuroimaging research, which often relies on understanding the hemodynamic response across modalities like fMRI and fNIRS. While fMRI measures the Blood-Oxygen-Level-Dependent (BOLD) signal, which is predominantly sensitive to deoxyhemoglobin, fNIRS uniquely quantifies both HbO and HbR concentrations simultaneously [45] [44]. This capacity positions fNIRS as a versatile tool for advancing our understanding of neurovascular physiology in both healthy and clinical populations, from studying the acute effects of concussion to monitoring long-term neurodegenerative processes [8] [46].
Core Biophysical Principles of fNIRS
Light-Tissue Interaction and the Modified Beer-Lambert Law
fNIRS operates on the principle that biological tissues are relatively transparent to light in the near-infrared spectrum (650-1000 nm). Within this "optical window," light can penetrate the scalp, skull, and brain tissue to a depth of several centimeters. The primary chromophores absorbing this light are oxyhemoglobin (HbO) and deoxyhemoglobin (HbR), which have distinct absorption spectra [45] [44]. A typical fNIRS system emits light at two or more specific wavelengths chosen to exploit the differential absorption properties of HbO and HbR.
The modified Beer-Lambert law (MBLL) forms the mathematical foundation for converting detected light attenuation into concentration changes of these chromophores. The MBLL relates the attenuation of light to the concentration of absorbers in a scattering medium, factoring in the increased path length due to scattering. The fundamental equations are [47]:
ΔOD = log10(I0/I) = ελ,HbO * Δ[HbO] * L * DPF + G
Where:
ΔODis the change in optical densityI0andIare the incident and detected light intensities, respectivelyελ,HbOandελ,HbRare the molar extinction coefficients of HbO and HbR at wavelength (λ)Δ[HbO]andΔ[HbR]are the changes in concentration of HbO and HbRLis the physical distance between source and detectorDPFis the differential pathlength factor, accounting for increased light path due to scatteringGis a geometric factor related to tissue scattering
By measuring attenuation at a minimum of two wavelengths, a system of equations is created and solved to compute the relative changes in HbO and HbR concentrations [47].
The Hemodynamic Response Function
The typical hemodynamic response to neural activation follows a characteristic pattern. Upon neuronal firing, a complex cascade of neurovascular coupling events leads to a localized increase in cerebral blood flow that exceeds the oxygen metabolic demand. This results in a predictable change in hemoglobin concentrations [32]:
- A increase in HbO due to the influx of oxygenated blood
- A decrease in HbR as oxygen is extracted at a lower rate than the blood flow increase
- A increase in total hemoglobin (HbT), reflecting the overall increase in cerebral blood volume
This hemodynamic response function (HRF) typically begins after a ~2-second delay post-stimulus, peaks around 6-10 seconds, and then gradually returns to baseline, sometimes followed by a slight undershoot [32]. The precise timing and shape of the HRF can vary based on brain region, age, and physiological conditions.
Diagram Title: Neurovascular Coupling Pathway
Experimental Protocols for fNIRS Research
Standardized Neurovascular Coupling Assessment
Robust experimental design is crucial for valid fNIRS measurements. A common approach involves block designs where periods of task performance alternate with rest or baseline conditions. The following protocol, adapted from studies on sport-related concussion, provides a framework for assessing neurovascular coupling [8]:
Pre-Test Preparation:
- Participants should abstain from caffeine, alcohol, and strenuous exercise for 12-24 hours before testing to minimize systemic physiological confounds.
- Testing should occur in a temperature-controlled room (e.g., 22.5°C) with minimal distractions.
- The fNIRS headpiece is positioned precisely, typically 1 cm above the eyebrow on the supraorbital ridge to avoid sinuses, ensuring optodes cover regions of interest like the dorsolateral prefrontal cortex (DLPFC).
Baseline Recording:
- Participants sit quietly for 5 minutes with eyes open, focused straight ahead, breathing normally.
- Researchers should avoid communication during this baseline period to maintain consistent physiological state.
Task Paradigm (Modified "Where's Wally/Waldo"):
- Implementation of a cognitive activation task such as the "Where's Wally" paradigm.
- The task consists of 5 cycles, each comprising:
- 20 seconds of eyes closed
- 40 seconds of eyes open actively searching for the target
- If the participant finds the target within 40 seconds, the slide advances immediately to maintain cognitive engagement.
- This design effectively elicits hemodynamic responses in the prefrontal cortex while being tolerable for various populations.
Data Acquisition Parameters:
- Sampling rate: Typically ≥ 10 Hz to capture physiological fluctuations
- Source-detector distance: 3-4 cm to ensure adequate cortical penetration
- Wavelengths: Commonly 760 nm and 850 nm to optimize differential absorption sensitivity
Multimodal Integration with EEG
Simultaneous fNIRS-EEG recording provides a comprehensive assessment of neurovascular function by combining hemodynamic and electrophysiological measures. A representative protocol for studying motor cortex activation illustrates this approach [48]:
Equipment Setup:
- EEG system with 32-electrode cap arranged according to the 10-10 international system
- fNIRS system with 8 optodes positioned over the sensorimotor cortex bilaterally
- Careful positioning to prevent physical interference between EEG electrodes and fNIRS optodes
Motor Task Paradigm:
- Participants perform self-paced right finger movements (e.g., sequential finger tapping) for a designated period
- Movement periods alternate with rest periods in a block design (e.g., 20s task/30s rest, repeated 10-15 times)
- Instructions emphasize consistent movement pace and minimal head motion
Synchronization:
- Precise temporal synchronization of fNIRS and EEG data streams using hardware triggers or software markers
- Recording of event markers indicating movement onset/offset for subsequent analysis
Data Analysis:
- For EEG: Calculation of event-related desynchronization (ERD) in alpha (8-13 Hz) and beta (13-30 Hz) frequency bands
- For fNIRS: Analysis of HbO and HbR concentration changes during movement periods
- Temporal correlation analysis to quantify neurovascular coupling by examining timing relationships between ERD and hemodynamic responses
Quantitative Relationships and Data Interpretation
Characteristic Hemodynamic Response Patterns
The table below summarizes typical fNIRS responses across different experimental paradigms, based on published research:
Table 1: Characteristic fNIRS Responses in Different Experimental Paradigms
| Experimental Paradigm | HbO Response | HbR Response | Brain Region | Temporal Characteristics | Key Correlations |
|---|---|---|---|---|---|
| Cognitive Task ("Where's Wally") [8] | Increase | Decrease | Prefrontal Cortex | Peak at 5-8s post-stimulus | Coupled with task performance |
| Motor Execution (Finger Tapping) [48] | Increase (Contralateral) | Decrease (Contralateral) | Sensorimotor Cortex | Onset ~2s, peak ~6-10s | Negative correlation with alpha/beta ERD (r² = -0.69/-0.54) |
| Auditory Stimulation (Intensity-Dependent) [32] | Increase with intensity | Decrease with intensity | Auditory Cortex | Dependent on stimulus parameters | Correlated with N1/P2 ERP amplitudes |
| Retired Athletes with mTBI [8] | Blunted/Reduced | Elevated/Reduced decrease | Prefrontal Cortex | Altered time-to-peak | Associated with concussion history |
Neurovascular Coupling Metrics
The relationship between electrophysiological activity and hemodynamic responses can be quantified through specific metrics, as demonstrated in simultaneous EEG-fNIRS studies:
Table 2: Temporal and Correlation Metrics in Neurovascular Coupling
| Parameter | Typical Value/Range | Interpretation | Experimental Context |
|---|---|---|---|
| HbO-ERD Delay | ~2.8s average [48] | Time between electrophysiological activation and hemodynamic response | Motor execution |
| HbO-Alpha Correlation | r² = -0.69 (p<0.0001) [48] | Strong negative correlation between HbO and alpha power | Motor execution |
| HbO-Beta Correlation | r² = -0.54 (p<0.0001) [48] | Negative correlation between HbO and beta power | Motor execution |
| HbR-Alpha Correlation | r² = +0.5 [48] | Positive correlation between HbR and alpha power | Motor execution |
| N1-HbR Correlation | Significant in left auditory cortex [32] | Relationship between auditory ERP and hemodynamic response | Auditory intensity processing |
| P2-HbR Correlation | Significant in right DLPFC [32] | Relationship between later ERP component and HbR | Auditory intensity processing |
Comparative Analysis with fMRI
Methodological and Technical Comparisons
Understanding the relative strengths and limitations of fNIRS and fMRI is essential for selecting the appropriate neuroimaging tool for specific research questions:
Table 3: Technical Comparison of fNIRS and fMRI
| Characteristic | fNIRS | fMRI |
|---|---|---|
| Primary Signal | HbO and HbR concentration changes | BOLD (primarily HbR-sensitive) |
| Spatial Resolution | Limited to cortical surfaces (~2-3 cm depth) [45] | Whole-brain, high resolution (~1-3 mm) [45] |
| Temporal Resolution | High (up to 100 Hz) [45] | Moderate (0.5-2 Hz) [45] |
| Portability | Fully portable/wireless systems available [45] | Stationary, requires MRI scanner |
| Participant Motion Tolerance | High tolerance for movement [45] | Highly motion-sensitive [45] |
| Environment | Naturalistic settings, bedside use [45] | Restricted to scanner environment [45] |
| Participant Population | Suitable for infants, children, clinical populations [45] | Limited for claustrophobic patients, individuals with metal implants [45] |
| Cost | Relatively affordable, one-time investment [45] | Expensive, high operational costs [45] |
| Physiological Basis | Direct measurement of HbO and HbR [44] | Indirect measurement based on magnetic properties of HbR [44] |
Complementary Strength in Multimodal Imaging
The combination of fNIRS and fMRI leverages their complementary strengths, providing a more comprehensive understanding of brain function [44]. Simultaneous fNIRS-fMRI recordings have demonstrated:
- Strong spatial correlation between fNIRS HbO signals and fMRI BOLD responses in motor tasks [49]
- The ability to disentangle vascular and metabolic components of the hemodynamic response through simultaneous measurement of HbO and HbR with fNIRS alongside BOLD fMRI
- Improved ecological validity through fNIRS measurements in naturalistic settings, with validation from fMRI findings obtained in controlled environments
Diagram Title: fMRI vs fNIRS Selection Workflow
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 4: Essential Materials and Equipment for fNIRS Research
| Item Category | Specific Examples | Function/Purpose |
|---|---|---|
| fNIRS Hardware Systems | OctaMon (Artinis); NIRSport, NIRScout (NIRx) [8] [49] | Base instrumentation for signal acquisition; varies in channel count and portability |
| Optodes | MRI-compatible optodes; Light-emitting and detecting optodes [49] | Interface with subject's head; sources emit NIR light, detectors capture reflected light |
| Wavelength Selection | 760 nm and 850 nm LEDs/lasers [45] | Optimize differential absorption between HbO and HbR |
| Headgear and Positioning | Dense array caps; 3D digitizers [45] | Secure optode placement; ensure consistent positioning and co-registration with anatomy |
| Data Analysis Software | AtlasViewer; Homer2; NIRS-SPM; Custom MATLAB/Python tools [45] [46] | Preprocessing, statistical analysis, and visualization of fNIRS data |
| Motion Correction Tools | Accelerometer-based systems; Algorithmic correction (e.g., PCA, wavelet) [46] | Identify and mitigate motion artifacts in signal |
| Physiological Monitoring | Pulse oximeter; Respiration belt; EEG system [48] [32] | Monitor systemic physiological confounds; enable multimodal integration |
| Calibration Standards | Optical phantoms with known absorption/scattering properties | Validate system performance and signal quality |
Decoding oxyhemoglobin and deoxyhemoglobin concentrations through fNIRS provides a critical window into the neurovascular system's functioning. The precise interpretation of these signals rests on a solid understanding of their biophysical foundations, their relationship to underlying neural activity through neurovascular coupling, and the methodological considerations involved in their acquisition. As the field advances, standardized protocols like those outlined here will enhance reproducibility and clinical translation.
The integration of fNIRS with other modalities, particularly EEG and fMRI, creates a powerful multimodal framework that overcomes the limitations of any single technique. This approach is especially valuable in clinical contexts such as monitoring treatment response in psychiatry [46] or detecting neurovascular abnormalities in neurological conditions [8]. Future developments in high-density arrays, wearable technology, and advanced analytical methods will further solidify fNIRS's role as an indispensable tool for decoding the complex relationship between brain activity, metabolism, and blood flow.
This technical guide provides a comprehensive overview of key experimental paradigms used in neurovascular coupling research, with a specific focus on functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS) methodologies. Neurovascular coupling (NVC) describes the fundamental physiological process whereby neural activity triggers localized changes in cerebral blood flow, a relationship that forms the basis for interpreting signals in both fMRI and fNIRS studies [32] [8]. Understanding the distinct experimental designs used to probe this relationship is crucial for researchers and drug development professionals aiming to investigate brain function, assess therapeutic efficacy, or develop novel diagnostics for neurological conditions.
The following sections detail specific paradigms, categorized by domain, and summarize their implementation, neural correlates, and associated hemodynamic responses. The integration of these techniques allows for a multi-faceted investigation of brain function, combining high temporal resolution from electrophysiology with the spatial information and hemodynamic tracking provided by fNIRS and fMRI.
Sensory Stimulation Paradigms
Sensory paradigms use controlled external stimuli to evoke reliable neural and hemodynamic responses, making them ideal for fundamental studies of neurovascular coupling.
Auditory Intensity Dependence
The Intensity-dependent amplitude changes (IDAP) paradigm investigates how the magnitude of brain responses scales with the physical intensity of an auditory stimulus, providing a window into basic sensory processing and neurovascular coupling efficiency [32].
- Experimental Protocol: In a typical fNIRS-EEG co-recording study, participants are presented with pure tone stimuli of varying intensities (e.g., 77.9 dB, 84.5 dB, and 89.5 dB). Each tone lasts for 500 ms and is presented in a random order multiple times (e.g., 54 times per intensity) to obtain a robust average response. Throughout the experiment, EEG records event-related potentials (ERPs) like the N1 and P2 components, while fNIRS monitors concurrent changes in oxyhemoglobin (O2Hb) and deoxyhemoglobin (HHb) concentrations in the auditory and prefrontal cortices [32].
- Key Findings: Studies using this paradigm consistently show an increase in N1, P2, and N1–P2 peak-to-peak ERP amplitudes with increasing sound intensity. In parallel, fNIRS data reveal a corresponding increase in O2Hb and a decrease in HHb in the auditory and prefrontal cortices. The correlation between the electrical (EEG) and hemodynamic (fNIRS) responses, particularly in the left auditory cortex for N1 and the right dorsolateral cortex for P2, provides strong evidence for neurovascular coupling [32].
Table 1: Summary of Key Findings in Auditory Intensity Paradigms
| Measurement Technique | Parameter | Response to Increased Intensity | Primary Brain Regions Involved |
|---|---|---|---|
| Electroencephalography (EEG) | N1 Amplitude | Increase | Auditory Cortex |
| Electroencephalography (EEG) | P2 Amplitude | Increase | Auditory Cortex |
| Functional NIRS (fNIRS) | Oxyhemoglobin (O2Hb) | Increase | Auditory Cortex, Prefrontal Cortex |
| Functional NIRS (fNIRS) | Deoxyhemoglobin (HHb) | Decrease | Auditory Cortex, Prefrontal Cortex |
Visual Cognitive Challenge (The "Where's Wally" Paradigm)
The "Where's Wally" (or "Where's Waldo") task is a validated visual cognitive challenge used to assess higher-order cognitive function and neurovascular coupling in a more ecologically valid setting [8].
- Experimental Protocol: Participants are seated in front of a screen and undergo a baseline period of 5 minutes of quiet rest. Following this, they perform the "Where's Wally" task, which typically consists of five cycles. Each cycle includes 20 seconds with eyes closed, followed by 40 seconds with eyes open, during which the participant actively searches for the character "Wally" in a complex visual scene. If found quickly, the image advances to a new one. The fNIRS headpiece is positioned over the prefrontal cortex to monitor hemodynamic changes throughout the task [8].
- Key Findings: This paradigm reliably elicits increased metabolic demand in the prefrontal cortex. In healthy controls, this is observed as a relative increase in O2Hb and a decrease in HHb. Studies on retired athletes with a history of mild traumatic brain injury (mTBI) have shown a statistically significant reduction in this hemodynamic response (specifically a blunted O2Hb increase in the left middle frontal gyrus) compared to controls, suggesting altered neurovascular coupling and cerebral metabolic demands following repeated head injuries [8].
Cognitive-Motor Task Paradigms
Cognitive-motor paradigms investigate the integration of planning, learning, and execution of movements, engaging a more distributed network of brain regions.
Rhythmic Auditory Stimulation
This paradigm leverages the natural connection between the auditory and motor systems to improve motor timing and performance, with significant applications in rehabilitation [50].
- Experimental Protocol: During gait or finger-tapping training, participants synchronize their movements to a repetitive auditory cue. This cue is often presented at a frequency slightly higher than the participant's baseline (e.g., preferred walking cadence). Motor performance, such as step timing, length, and variability, is quantified kinematically. Neuroimaging during such tasks often involves fMRI or fNIRS to capture associated activation in motor and timing-related brain circuits [50].
- Key Findings: Research demonstrates that rhythmic auditory cues can immediately improve gait kinematics in patients with Parkinson's disease and post-stroke, increasing speed, stride length, and reducing variability. Neuroimaging studies link this behavioral improvement to the activation of a network including the supplementary motor area (SMA), premotor cortex, superior temporal gyrus, prefrontal cortex, and cerebellum. The rich structural connectivity between auditory and motor regions, including projections from auditory areas to the basal ganglia and cerebellum, which in turn project to the SMA and premotor cortex, underpins this effective sensorimotor integration [50].
Grid-Sailing Task (GST) for Internally-Guided Sequencing
The Grid-Sailing Task (GST) is a canonical paradigm for studying internally-guided motor skills, where the sequence of actions is self-generated rather than dictated by external cues [51].
- Experimental Protocol: Participants navigate a cursor from a start position to a goal position on a 5x5 grid using a specific key-mapping (KM) that associates keyboard buttons with cursor movement directions. The instruction is to reach the goal in the fewest steps as quickly as possible. To dissect learning components, participants may train on a single start-goal (SG) configuration repeatedly (Single-SG condition, promoting sequence-specific motor learning) or on multiple novel SG configurations using the same KM (Mixed-SG condition, promoting the acquisition of a KM-specific internal model, or cognitive learning) [51].
- Key Findings: The GST has been instrumental in disentangling the contributions of cognitive and motor learning. Performance improvements in the Single-SG condition are attributed to motor learning of the specific movement trajectory. In contrast, improvements and transfer of skills to novel SG configurations in the Mixed-SG condition provide evidence for cognitive learning—the acquisition of a flexible, internal model of the key-mapping that facilitates efficient trajectory planning. This distinguishes it from externally-specified sequencing tasks like the Serial Reaction Time (SRT) task, where movements are directly cued by stimuli [51].
Table 2: Comparison of Externally-Specified vs. Internally-Guided Sequencing Paradigms
| Feature | Externally-Specified Sequencing (e.g., SRT Task) | Internally-Guided Sequencing (e.g., GST) |
|---|---|---|
| Sequence Source | Environment provides sequence of stimuli | Self-generated, internally specified |
| Action Control | Stimulus-Response (S-R) bindings | Anticipated Action Effects (R-E bindings) |
| Learning Type | Sensorimotor Learning | Ideomotor Learning |
| Primary Neural Correlates | Cerebellar, Premotor circuits [51] | Basal Ganglia, Pre-SMA, DLPFC [51] |
| Example Tasks | Serial Reaction Time, Discrete Sequence Production | Drawing, Composing music, Grid Navigation |
The following diagram illustrates the workflow for a typical multi-modal neurovascular coupling study, integrating the paradigms discussed above:
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful execution of the described paradigms requires a suite of specialized equipment and software. The following table details key components of a neurovascular coupling research toolkit.
Table 3: Essential Research Reagents and Solutions for NVC Studies
| Item Name | Function/Application | Specific Examples/Notes |
|---|---|---|
| fNIRS System | Measures hemodynamic changes by detecting absorption of near-infrared light by O2Hb and HHb. | Multi-channel systems (e.g., OctaMon); used to probe auditory, prefrontal, and motor cortices [32] [8]. |
| EEG System | Records millisecond-resolution electrical brain activity from the scalp. | Critical for capturing event-related potentials (ERPs) like N1/P2 in auditory paradigms [32]. |
| fMRI Scanner | Measures brain activity via the Blood-Oxygen-Level-Dependent (BOLD) signal. | Provides high spatial resolution; less suitable for noisy auditory paradigms [32]. |
| Stimulus Presentation Software | Precisely controls the timing and delivery of sensory and cognitive tasks. | Used to present auditory tones, visual "Where's Wally" tasks, and Grid-Sailing Task displays [32] [8] [51]. |
| Rhythmic Auditory Stimulator | Generates precise rhythmic cues for motor synchronization. | Metronomes or specialized software for gait and finger-tapping studies [50]. |
| Key-Mapping Interface | Translates participant keypresses into actions within a virtual task. | Essential for implementing the Grid-Sailing Task and other internally-guided paradigms [51]. |
The core physiological process of neurovascular coupling, which is measured using the tools above, can be summarized as follows:
Neurovascular coupling (NVC) represents the critical physiological process that dynamically links neuronal activity to subsequent changes in local cerebral blood flow and oxygenation. This whitepaper examines how NVC dysfunction serves as a fundamental mechanism and biomarker across three major neurological conditions: Alzheimer's disease (AD), cerebrovascular disease, and mild traumatic brain injury (mTBI). Research over the past three decades has established that impaired NVC mechanisms contribute significantly to disease pathogenesis and progression, offering promising avenues for early diagnosis and therapeutic intervention. The integration of advanced neuroimaging techniques, particularly functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS), has enabled researchers to quantify these impairments with increasing precision. This technical guide provides a comprehensive overview of current research applications, experimental protocols, and analytical frameworks for investigating NVC in disease contexts, specifically tailored for researchers, scientists, and drug development professionals working in neuroscience.
Neurovascular coupling constitutes a complex biological process essential for maintaining brain homeostasis by ensuring that active brain regions receive adequate blood supply to meet metabolic demands. The functional unit responsible for this process, the neurovascular unit (NVU), comprises neurons, astrocytes, vascular smooth muscle cells, pericytes, and endothelial cells that work in concert to regulate cerebral blood flow [7]. Over the past 30 years, research into NVC has expanded exponentially, with the United States maintaining a clear leading position in output, while China has demonstrated rapid growth in research participation over the past decade [7] [39].
In pathological conditions, neurovascular uncoupling disrupts the precise temporal and spatial coordination between neural activity and hemodynamic responses. This dysfunction has been implicated as one of the earliest detectable abnormalities in multiple neurological disorders, often preceding overt structural damage or clinical symptom manifestation [13] [1]. The study of NVC impairments provides valuable insights into disease mechanisms and offers a window into the functional integrity of the cerebral microvasculature. Keyword analysis of the NVC research landscape has identified "cerebral blood flow," "neuronal activity," and "neurovascular coupling" as dominant terms, emphasizing the central role of brain function and imaging techniques in this field [7].
Table 1: Neurovascular Coupling Research Metrics (1996-2025)
| Metric | Value | Significance |
|---|---|---|
| Total Publications | 2,047 articles | Steady growth over 30-year period |
| Annual Growth Rate | 16.5% | Rapidly expanding field |
| Citation Peak | Nearly 10,000 (2022-2023) | High research impact |
| Leading Research Country | United States (663 articles, 32.4%) | Clear leadership position |
| Emerging Research Country | China (195 articles, 9.5%) | Rapid growth over past decade |
Neurovascular Coupling Mechanisms and Dysfunction in Disease
Molecular Mechanisms of NVC
The signaling pathways governing NVC involve sophisticated interplay between multiple vasoactive compounds released by neurons and astrocytes in response to increased neural activity. Under healthy conditions, neuronal depolarization leads to calcium influx, activating neuronal nitric oxide synthase (nNOS) and generating nitric oxide (NO), a potent vasodilator [13]. Systematic reviews of in vivo experiments indicate that pharmacological or genetic knockout of nNOS causes the largest reduction in neurovascular response, with an average decrease of 64% across 11 studies [13]. NO mediates vasodilation primarily through raising cyclic guanosine monophosphate (cGMP) levels in vascular smooth muscle cells, though interestingly, this mechanism does not appear to function similarly in pericytes [13].
Additional pathways involve the metabolism of arachidonic acid (AA) into vasoactive agents. Rather than being primarily mediated by phospholipase A2 (PLA2) as previously thought, recent evidence identifies phospholipase D2 (PLD2) as the initiator of AA synthesis in NVC [13]. AA is subsequently metabolized into vasodilatory species including prostaglandin E2 (PGE2) and various epoxyeicosatrienoic acids (EETs), which act on the prostaglandin E2 receptor 4 (EP4) receptor on capillary pericytes and vascular smooth muscle cells [13]. The EP4 receptor functions as a Gs-linked G-protein-coupled receptor (GPCR), whose activation increases intracellular cAMP, phosphorylates myosin light chain kinase, and ultimately leads to vasodilation [13].
Purinergic signaling represents another important mechanism, with astrocyte calcium signals playing a necessary role in capillary dilation via pericytes, though curiously not in arteriolar dilation by vascular smooth muscle cells [13]. The precise initiator of this astrocytic calcium signal remains controversial, with evidence challenging the traditional model of astrocytic metabolic glutamate receptor 5 (mGluR5) activation by synaptic glutamate release [13].
Disease-Specific NVC Dysfunction
Alzheimer's Disease
In Alzheimer's disease, NVC dysfunction emerges as one of the earliest reliable biomarkers, potentially preceding and even paving the way for amyloid-β (Aβ) pathology [13]. The relationship between cerebrovascular dysfunction and AD is bidirectional, with evidence suggesting that NVC impairment may facilitate Aβ accumulation, while Aβ peptides themselves exert significant neurovascular effects that contribute to cognitive decline and disease progression [13]. Amyloid-β deposition adversely affects endothelial function and pericyte signaling, thereby compromising the NVU's ability to match blood flow to neural demand [7] [13]. Animal models of AD demonstrate impaired functional hyperemia long before the appearance of amyloid plaques, and these abnormalities can be reproduced in normal mice with superficial application of amyloid-beta peptide (Aβ1-40) on the neocortex [1].
Metabolic Syndrome and Cerebrovascular Disease
The metabolic syndrome represents a cluster of conditions that significantly impact cerebrovascular health and NVC function. Recent research utilizing fNIRS has demonstrated that older adults with metabolic syndrome exhibit impaired NVC during higher cognitive loads, as reflected by an attenuated increase in oxyhemoglobin (HbO) in the premotor cortex during demanding tasks [52]. These impairments correlate with poorer peripheral endothelial function, as measured by flow-mediated dilation (FMD), and reduced accuracy on cognitive tasks, suggesting that peripheral vascular dysfunction may be implicated in cerebrovascular dysregulation [52]. This NVC impairment likely represents an early cerebrovascular mechanism underpinning cognitive decline in metabolic syndrome, discernible particularly during higher cognitive loads [52].
Mild Traumatic Brain Injury
In mild traumatic brain injury, particularly sports-related concussion, NVC disruption represents a persistent consequence of head trauma. Research involving retired rugby players with histories of multiple concussions has demonstrated significantly reduced cerebral hemodynamic responses compared to controls without concussion history [53] [8]. These impairments manifest as altered oxygen extraction patterns, with the mTBI group showing a greater rate of oxygen extraction compared to controls, suggesting altered cerebral metabolic demands following repeated head injuries [53]. The pathophysiology involves endothelial cell impairment of the blood-brain barrier, mitochondrial dysfunction, and disruption of cerebral autoregulation and cerebral blood flow velocity [53].
Table 2: NVC Dysfunction Across Neurological Conditions
| Disease | Primary NVC Defect | Key Molecular Mechanisms | Functional Consequences |
|---|---|---|---|
| Alzheimer's Disease | Impaired functional hyperemia | Amyloid-β effects on endothelial function and pericyte signaling; disrupted NO signaling | Early cognitive decline; disrupted cerebral blood flow regulation |
| Metabolic Syndrome | Attenuated HbO response during cognitive load | Peripheral endothelial dysfunction; reduced flow-mediated dilation | Working memory deficits; accelerated cognitive decline |
| mTBI | Reduced hemodynamic response; altered O2 extraction | Endothelial impairment; mitochondrial dysfunction; autoregulation disruption | Cognitive impairment; altered metabolic demands |
Experimental Protocols and Methodologies
fNIRS Protocols for NVC Assessment
Functional near-infrared spectroscopy (fNIRS) has emerged as a particularly valuable tool for assessing NVC in clinical populations due to its non-invasive nature, portability, and relative tolerance to motion artifacts. A standardized protocol for NVC assessment using fNIRS involves the following components:
Participant Preparation: Participants should be tested in a temperature-controlled environment (approximately 22.5°C) without distractions. They should be hydrated and refrain from exercise, caffeine (for 12 hours), and alcohol (for 24 hours) prior to testing [53]. Basic anthropometric measurements including height and weight should be recorded.
fNIRS Device Setup: The fNIRS cranial headpiece is typically placed 1 cm above the eyebrow on the supraorbital ridge to avoid the sinuses. An 8-channel system can monitor changes in oxyhemoglobin (O2Hb) and deoxyhemoglobin (HHb) in prefrontal regions, covering the dorsolateral (DLPFC) and orbitofrontal (OFC) cortices in both hemispheres [53].
The Modified Neary Protocol: This well-validated approach for NVC assessment utilizes cognitive activation to elicit measurable hemodynamic responses [53] [8]. The protocol consists of:
- Baseline Period: Participant sits quietly for 5 minutes with eyes open, focused straight ahead.
- Cognitive Activation: Implementation of the "Where's Wally" (or "Where's Waldo") paradigm, consisting of five cycles (5 minutes total), each with 20 seconds of eyes closed followed by 40 seconds of eyes open while visually searching for the target character.
- Data Acquisition: Continuous measurement of O2Hb and HHb concentration changes throughout the protocol.
Outcome Measures: Key parameters include relative changes in O2Hb and HHb concentrations, total hemoglobin (tHb = O2Hb + HHb) as an indicator of blood volume, and hemoglobin difference (HbDiff = O2Hb - HHb) reflecting oxygen extraction and metabolic cellular changes [53].
fMRI Protocols for NVC Assessment
Functional magnetic resonance imaging (fMRI) provides superior spatial resolution for investigating NVC mechanisms, with blood oxygen level-dependent (BOLD) fMRI being the most widely used approach. The BOLD signal indicates active brain regions based on the tight correlation between neuronal activity and increased blood flow, which causes a rapid decrease in paramagnetic deoxyhemoglobin detectable via MRI [13].
Data Acquisition Parameters:
- Spatial resolution: Typically 1-2 mm isotropic voxels
- Temporal resolution: Approximately 1 second
- Task design: Block or event-related paradigms tailored to specific cognitive domains
- Complementary techniques: Arterial spin labeling (ASL) for direct cerebral blood flow measurement
Preprocessing Pipeline:
- Motion Correction: Modeling and removal of motion parameters, their derivatives, and quadratics (24 total nuisance parameters) [54]
- Physiological Noise Removal: aCompCor method for ventricle and white matter time series using first five principal components for each plus derivatives and quadratics (40 total nuisance parameters) [54]
- Spatial Normalization: Alignment to standard template space
- Spatial Smoothing: Typically with a Gaussian kernel (though note spatial autocorrelation considerations for activity flow mapping)
Activity Flow Mapping: This innovative approach models the movement of task-evoked activity over brain connections to predict task-evoked activations [54]. The methodology involves:
- Formatting task activation data as nodes by task conditions by subjects
- Estimating functional connectivity from resting-state or task-state data
- Applying the Brain Activity Flow Toolbox to simulate activity flow processes
- Validating predictions against empirically observed task-evoked activity
Computational Modeling and Analysis Frameworks
Dynamic Models of NVC
Computational models for interpreting NVC data incorporate biophysical parameters related to neurovascular coupling, with seven major models identified in the literature exhibiting strong differences in complexity [1]. These models generally include multiple compartments, each modeling a particular step in the global process from neural activity to observed signal. The Balloon model and Windkessel model are based on hypothetical inflation or deflation of cerebral venules secondary to variations of intravascular pressure with inflow [1].
These dynamic models serve two primary purposes: (1) as descriptive models for depicting transient hemodynamic and oxygenation changes in activated cerebral areas, and (2) as explanatory models that mimic physiological mechanisms and estimate various biophysical parameters related to fMRI signal changes [1]. When applied to pathological conditions, these models can extract key physiological information from signal changes in functional imaging studies, helping to interpret the underlying NVC mechanisms disrupted in disease states [1].
Hybrid Decomposition Frameworks
The NeuroMark pipeline represents an advanced hybrid approach for functional decomposition of neuroimaging data, combining elements of both predefined atlases and data-driven solutions [55]. This framework categorizes functional decompositions along three primary attributes:
- Source: Anatomic, functional, or multimodal
- Mode: Categorical (discrete regions) or dimensional (continuous, overlapping representations)
- Fit: Predefined, data-driven, or hybrid
The NeuroMark approach uses a template derived from running blind independent component analysis (ICA) on multiple large datasets to identify a replicable set of components, which are then used as spatial priors in a single-subject spatially constrained ICA analysis [55]. This allows estimation of subject-specific maps and timecourses while maintaining correspondence between individuals, facilitating cross-subject comparisons essential for clinical research.
Signaling Pathways and Visualization
The molecular mechanisms underlying neurovascular coupling involve complex interactions between multiple cell types and signaling pathways. The following diagram illustrates the primary signaling cascades involved in neurovascular coupling and how they become disrupted in disease states:
Neurovascular Coupling Signaling Pathways in Health and Disease
The experimental workflow for assessing NVC function in neurological disorders follows a systematic process from participant recruitment to data interpretation, as illustrated in the following diagram:
Experimental Workflow for NVC Assessment in Neurological Disorders
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Materials and Analytical Tools for NVC Investigation
| Tool/Category | Specific Examples | Function/Application | Technical Notes |
|---|---|---|---|
| Neuroimaging Platforms | fNIRS systems (OctaMon, Artinis); fMRI scanners (3T, 7T) | Hemodynamic response measurement; brain activation mapping | fNIRS: prefrontal cortex coverage; fMRI: whole-brain BOLD signal |
| Cognitive Paradigms | "Where's Wally" task; n-back task; intensity-dependent auditory stimuli | NVC activation during controlled cognitive load | Parametric intensity variation enhances signal detection |
| Computational Toolboxes | Brain Activity Flow Toolbox; NeuroMark pipeline; bibliometrix R package | Activity flow mapping; functional decomposition; scientometric analysis | Python/NumPy compatibility; spatial autocorrelation correction |
| Physiological Monitors | Brachial ultrasound (FMD); heart rate/respiration monitors | Peripheral endothelial function; physiological artifact control | aCompCor for noise removal in fMRI |
| Analytical Frameworks | Balloon model; Windkessel model; general linear model (GLM) | Hemodynamic response modeling; activation statistical analysis | Finite impulse response (FIR) for task-state FC estimation |
| Spatial Decomposition | Independent component analysis (ICA); atlas-based parcellations (AAL, Yeo) | Functional network identification; region-of-interest definition | Hybrid approaches balance individual variability with cross-study comparison |
The investigation of neurovascular coupling in neurological diseases has evolved from a specialized research area to a central focus in understanding disease mechanisms and developing biomarkers. The integration of multimodal imaging approaches, particularly fMRI and fNIRS, with advanced computational modeling has enabled unprecedented insights into the functional integrity of the neurovascular unit across pathological conditions. Research over the past three decades has established NVC dysfunction as an early event in Alzheimer's disease, a consequence of cerebrovascular risk factors in metabolic syndrome, and a persistent effect of mild traumatic brain injury.
Future research directions will likely focus on integrating artificial intelligence, multi-omics analysis, and high-resolution imaging to further elucidate NVC mechanisms in health and disease [7]. The development of dynamic fusion models that incorporate multiple time-resolved symmetric data fusion decompositions will enhance our ability to characterize complex brain dynamics [55]. Additionally, the standardization of assessment protocols and analytical pipelines, as demonstrated by initiatives like the NeuroMark framework, will improve reproducibility and clinical translation of NVC biomarkers. As these methodological advances continue to mature, assessment of neurovascular coupling is poised to become an essential component of both clinical trial design and therapeutic monitoring in neurological disorders.
Advantages and Limitations of Each Technique for Specific Research Scenarios
Functional neuroimaging has revolutionized our understanding of brain function by enabling non-invasive measurement of brain activity in living humans. Two prominent techniques—functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS)—have become indispensable tools for investigating neurovascular coupling, the fundamental physiological process that links neuronal activity to subsequent changes in cerebral blood flow and oxygenation [56] [12]. This technical guide provides a comprehensive comparison of fMRI and fNIRS, examining their respective advantages, limitations, and optimal applications within specific research scenarios, particularly for researchers and drug development professionals investigating brain function in health and disease.
The neurovascular coupling mechanism forms the physiological basis for both fMRI and fNIRS. When neurons become active, they trigger a complex cascade of cellular events involving astrocytes, pericytes, and endothelial cells that ultimately leads to localized vasodilation of arterioles and increased cerebral blood flow [56] [12]. This functional hyperemia delivers oxygen and glucose to meet the metabolic demands of active neurons while removing waste products. fMRI detects these changes primarily through the blood-oxygen-level-dependent (BOLD) signal, which reflects the magnetic susceptibility differences between oxygenated and deoxygenated hemoglobin [56] [57]. In contrast, fNIRS directly measures concentration changes in oxygenated (HbO) and deoxygenated hemoglobin (HbR) in cortical microvessels by exploiting the differential absorption properties of near-infrared light by these chromophores [32] [58] [57].
Technical Principles and Measurement Foundations
fMRI: Blood-Oxygen-Level-Dependent (BOLD) Contrast
Functional MRI relies on the BOLD contrast mechanism, which indirectly reflects neural activity through changes in blood oxygenation. During neuronal activation, a localized increase in cerebral blood flow delivers more oxygenated hemoglobin than is consumed, leading to a decrease in deoxygenated hemoglobin concentration [56] [57]. Since deoxyhemoglobin is paramagnetic (while oxyhemoglobin is diamagnetic), this change alters the magnetic susceptibility of the tissue, increasing the T2* relaxation time and resulting in an increased BOLD signal [57]. The BOLD signal represents a complex mixture of changes in cerebral blood flow, cerebral blood volume, and the cerebral metabolic rate of oxygen consumption [56]. A critical assumption underlying BOLD fMRI is that neurovascular coupling remains intact, which may be compromised in various pathological conditions including brain tumors, stroke, and neurodegenerative diseases [56].
fNIRS: Optical Absorption of Hemoglobin Species
Functional NIRS utilizes near-infrared light (650-950 nm) to penetrate biological tissues, including the skull, and measures changes in light absorption by the primary chromophores in brain tissue: oxygenated and deoxygenated hemoglobin [58] [57] [59]. Within the near-infrared spectrum, biological tissues are relatively transparent, creating an "optical window" that enables light to reach the cerebral cortex and be detected after passing through the brain [60] [59]. The technique is based on the modified Beer-Lambert law, which relates changes in light attenuation to changes in chromophore concentrations [61]. During neuronal activation, neurovascular coupling typically produces an increase in HbO and a decrease in HbR concentrations in the activated brain region due to an oversupply of oxygenated blood [32] [58]. This hemodynamic response function typically begins after a 2-second delay following neural activation, peaks at 6-10 seconds, and then gradually returns to baseline [32].
Table 1: Fundamental Measurement Characteristics of fMRI and fNIRS
| Characteristic | fMRI | fNIRS |
|---|---|---|
| Primary Measurement | BOLD signal (T2* changes) | Concentration changes of HbO and HbR |
| Physiological Basis | Magnetic susceptibility of deoxyhemoglobin | Optical absorption of hemoglobin species |
| Spatial Resolution | High (1-2 mm) [12] | Moderate (cortical surface only) [57] [61] |
| Temporal Resolution | Moderate (~1 second) [12] | High (up to 10 Hz typically, potentially higher) [57] |
| Penetration Depth | Whole brain | Superficial cortex (1-3 cm) [60] [61] |
| Directly Measured Parameters | BOLD signal ratio | Separate HbO and HbR concentrations [57] |
Comparative Analysis: Advantages and Limitations
Technical and Practical Considerations
Table 2: Advantages and Limitations of fMRI and fNIRS
| Aspect | fMRI | fNIRS |
|---|---|---|
| Key Advantages | - Excellent spatial resolution (1-2 mm) [12]- Whole-brain coverage including deep structures [57]- Well-established analysis pipelines- High anatomical specificity when combined with structural MRI | - High portability and cost-effectiveness [58] [57] [59]- Tolerance to movement artifacts [59]- Non-invasive and safe for repeated measurements [57]- Silent operation suitable for auditory studies [32]- Ability to measure during naturalistic activities [58] [59] |
| Principal Limitations | - Susceptibility to motion artifacts- High cost and limited accessibility- Loud acoustic noise interfering with auditory paradigms [32]- Confinement in scanner creates artificial environment- Not suitable for patients with metal implants or claustrophobia [57] | - Limited to cortical surface measurements [57] [61]- Lower spatial resolution than fMRI [57]- Signal contamination from extracerebral tissues (scalp, skull) [60]- Limited penetration depth (worse in individuals with dense skulls or dark hair) [58]- Lack of standardized analysis procedures [60] |
| Research Scenarios Best Suited | - Mapping entire functional networks- Studies requiring precise anatomical localization- Investigating deep brain structures- Clinical preoperative mapping [56] | - Studies requiring ecological validity [59]- Infant and child development research [59]- Social interaction studies [59]- Motor rehabilitation monitoring [59]- Long-duration monitoring [57]- Auditory processing research [32] |
Neurovascular Coupling Integrity in Pathological Conditions
Both fMRI and fNIRS rely on the assumption of intact neurovascular coupling, but this coupling can be disrupted in various pathological conditions. Brain tumors, particularly gliomas, can invade the perivascular space and disrupt normal interactions between astrocytes, pericytes, and endothelial cells, leading to neurovascular uncoupling (NVU) [56]. This uncoupling can confound the interpretation of fMRI data in presurgical mapping, potentially leading to neurological complications if not properly accounted for [56]. Similar disruptions have been observed in cerebrovascular diseases, neurodegenerative conditions including Alzheimer's and Parkinson's disease, and following stroke [56] [12].
In conditions where neurovascular coupling is compromised, fNIRS may offer some advantages due to its ability to separately measure HbO and HbR concentrations, allowing for more detailed analysis of hemodynamic responses [57]. Furthermore, the suppression of anomalous tumor blood vessel formation with antiangiogenic therapies has been shown to "normalize" brain tumor vasculature and potentially restore neurovascular coupling, which could be monitored using fNIRS [56].
Experimental Protocols for Neurovascular Coupling Assessment
Protocol 1: Multimodal Assessment of Neurovascular Coupling
A comprehensive approach to investigating neurovascular coupling involves simultaneous measurement using EEG, fNIRS, and transcranial Doppler ultrasound (TCD) [62]. This protocol enables direct correlation of neuronal activity (EEG) with microvascular oxygenation changes (fNIRS) and macrovascular flow velocity alterations (TCD).
Methodology:
- Participants: 15 healthy adults (age range 19-32)
- Experimental tasks: Motor (finger tapping) and visual ("Where's Waldo?" search task)
- Synchronized monitoring: Blood pressure, capnography, and heart rate throughout
- fNIRS placement: Frontal, motor, parietal, and occipital cortices
- TCD insonation: Middle and posterior cerebral arteries
- EEG setup: 16 channels according to the 10-20 system
- Analysis: Time-frequency analysis of EEG, cross-correlation between hemodynamic and neuronal responses
Key Findings: The visual task elicited robust responses across all modalities, with cross-correlation analysis demonstrating that changes in oxygenated hemoglobin and cerebral blood velocity had a moderate-to-strong negative correlation with systemic physiological influences, confirming that measured changes resulted from neuronal input rather than systemic factors [62].
Protocol 2: Clinical Application in Stroke Populations
fNIRS has emerged as a valuable tool for monitoring cortical reorganization and functional recovery following stroke, particularly due to its tolerance to movement and portability [59] [63].
Methodology:
- Participants: 107 individuals with history of left or right hemisphere stroke
- Experimental tasks: Resting state, discourse comprehension, and picture naming (varying cognitive and motor speech demands)
- fNIRS signal quality assessment: Scalp-coupling indices, peak spectral power values, and number of bad channels
- Quality assessment tool: Quality Testing of Near Infrared Scans (QT-NIRS) toolbox
- Analysis: Examination of protocol factors and participant characteristics on data quality
Key Findings: Data quality was not affected by session location or protocol experience but was significantly lower during picture naming compared to other tasks. Importantly, fNIRS signals were generally worse for Black women compared to Black men and White individuals regardless of gender, highlighting the need to address potential biases in optical instrumentation [63].
Visualization of Neurovascular Coupling and Measurement Techniques
Figure 1: Neurovascular Coupling and Measurement Techniques
Research Reagent Solutions and Essential Materials
Table 3: Essential Research Materials for Neurovascular Coupling Studies
| Item | Function/Application | Technical Notes |
|---|---|---|
| fNIRS Systems | Measurement of cortical hemodynamics | - Types: Continuous-wave (CW), frequency-domain (FD), time-domain (TD) [61]- CW systems: Most common, cost-effective, relative measurements [61]- FD systems: Absolute quantification, more complex [61] |
| fNIRS Caps | Standardized optode placement | - International 10-20 system locations [61]- Short-separation detectors (8mm) for superficial signal correction [61] |
| Analysis Software | Data processing and visualization | - HOMER3: MATLAB-based analysis toolbox [61]- NIRS Toolbox: Object-oriented MATLAB framework [61]- AtlasViewer: Probe design and brain visualization [61] |
| Multimodal Integration | Comprehensive NVC assessment | - EEG-fNIRS: Neuronal + hemodynamic correlation [62]- fNIRS-TCD: Microvascular + macrovascular assessment [62]- Physiological monitoring: Blood pressure, heart rate, capnography [62] |
The selection between fMRI and fNIRS for neurovascular coupling research depends critically on the specific research questions, participant population, and experimental context. fMRI remains the gold standard for high spatial resolution mapping of entire brain networks, including deep structures, making it ideal for preoperative mapping and studies requiring precise anatomical localization. In contrast, fNIRS offers unique advantages for ecological validity, naturalistic settings, longitudinal monitoring, and studies involving populations challenging to scan in traditional MRI environments, such as infants, children, elderly individuals, and patients with movement disorders.
Emerging approaches that combine multiple neuroimaging modalities show particular promise for comprehensive assessment of neurovascular coupling across different spatial and temporal scales. The integration of fNIRS with EEG, TCD, and other physiological monitoring techniques enables researchers to dissect the complex relationships between neuronal activity, microvascular oxygenation changes, and macrovascular flow dynamics. As both technologies continue to evolve, their complementary strengths will further enhance our understanding of neurovascular coupling in both healthy brain function and neurological disorders, ultimately advancing drug development and clinical care for patients with cerebrovascular and neurodegenerative conditions.
Troubleshooting and Optimization: Overcoming Technical and Physiological Confounds in NVC Studies
Functional near-infrared spectroscopy (fNIRS) and functional magnetic resonance imaging (fMRI) have become cornerstone techniques in cognitive neuroscience and clinical research for assessing brain function through neurovascular coupling (NVC)—the fundamental process linking neuronal activity to subsequent hemodynamic changes [32] [8]. However, the measured signals are notoriously susceptible to various confounding artifacts that can obscure genuine neural correlates, leading to both false positives and false negatives in research findings [64] [65]. For fNIRS, the measured signal comprises multiple components, including the neuronal-evoked changes of interest, systemic physiological changes in cerebral and extracerebral compartments, vascular evoked changes, and muscular activity [64]. This technical guide provides an in-depth examination of the primary artifact sources—physiological noise, motion, and systemic confounds—framed within the context of NVC research, and offers evidence-based methodologies for their mitigation, enabling more accurate interpretation of neuroimaging data.
Physiological Noise and Systemic Confounds
Origins and Impact on Signal Fidelity
Physiological noise originates from the body's inherent rhythms and regulatory mechanisms. The cardiorespiratory system (heartbeat ~1 Hz, respiration ~0.2-0.3 Hz), Mayer waves (~0.1 Hz), and spontaneous fluctuations in arterial blood pressure and CO₂ concentration constitute major noise sources [64] [66]. These systemic physiological activities are a significant challenge because they can mimic the hemodynamic response characteristic of neuronal activation, creating "false positive" results, or mask a true neuronal response, leading to "false negatives" [64]. For instance, an increase in arterial partial-pressure of CO₂ (PaCO₂) can produce a hemodynamic change that closely resembles a true task-related response, characterized by a large increase in oxyhemoglobin (HbO) and a slight decrease in deoxyhemoglobin (HbR) [64].
The complexity of properly interpreting fNIRS signals is further heightened by the fact that the contribution of each signal component varies based on experimental paradigm, measurement location, technical implementation, and individual participant physiology [64]. Crucially, extracerebral systemic physiology, primarily from the scalp, can dominate the fNIRS signal, with some estimates suggesting it contributes roughly 94% of the signal measured by a standard channel [67].
Table 1: Characteristics of Major Physiological Noise Sources
| Noise Source | Typical Frequency Range | Primary Origin | Impact on Hemoglobin Concentrations |
|---|---|---|---|
| Cardiac Pulsation | ~1 Hz [66] | Arterial pulsatility in cerebral and extracerebral tissues | Pseudo-periodic, high-frequency spikes in both HbO and HbR [66] |
| Respiration | ~0.2-0.3 Hz [66] | Changes in intrathoracic pressure affecting cerebral blood flow | Low-frequency oscillations in both HbO and HbR [66] |
| Mayer Waves | ~0.1 Hz [66] | Spontaneous oscillations in systemic blood pressure and sympathetic nerve activity | Low-frequency oscillations correlated with blood pressure changes [65] |
| CO₂ Fluctuations | Very Low Frequency (<0.05 Hz) | Changes in end-tidal CO₂ affecting cerebral vasodilation/constriction | Can mimic functional activation: increases HbO, decreases HbR [64] [65] |
Mitigation Strategies and Experimental Protocols
Several effective methodologies have been developed to combat physiological noise. A powerful approach is Systemic Physiology Augmented fNIRS (SPA-fNIRS), which involves the simultaneous recording of systemic physiological parameters—such as heart rate, mean arterial pressure, and end-tidal CO₂—to use as regressors in post-processing [64] [67]. This allows for the statistical removal of these confounding components from the fNIRS signal.
Another robust technique is the use of short-separation channels (SSS). These are fNIRS channels with a small source-detector distance (typically ~0.8 cm for adults) that are primarily sensitive to hemodynamic changes in the extracerebral layers [67]. The data from these channels can be incorporated into a General Linear Model (GLM) as nuisance regressors to subtract the superficial confounds from the standard long-separation channels [67].
Adaptive filtering techniques, such as Recursive Least-Squares Estimation (RLSE), have also been successfully employed. One study modeled the fNIRS signal as a linear combination of the expected hemodynamic response, short-separation data, physiological noises (modeled as a sum of three sinusoidal functions), and baseline drift [66]. This RLSE method demonstrated significant noise reduction, achieving a 77% and 99% improvement in the number of channels with higher contrast-to-noise ratio for HbO and HbR, respectively, compared to other methods [66].
Figure 1: Workflow for mitigating physiological noise in fNIRS, illustrating primary noise sources and methodological solutions.
Motion Artifacts
Characterization and Challenges
Motion artifacts are a pervasive source of noise in fNIRS data, especially in studies involving mobile participants, clinical populations, or infants [68]. These artifacts occur when head movement causes a temporary decoupling between the optical fibers and the scalp, resulting in abrupt, high-amplitude shifts in the measured intensity [68]. Motion artifacts can be categorized as spikes, baseline shifts, and low-frequency variations [68]. A particularly challenging type of artifact is one that is temporally correlated with the task, such as the low-frequency, low-amplitude motion induced by jaw movement during vocal responses, which can be difficult to distinguish from the true hemodynamic response [68].
The traditional approach of rejecting contaminated trials is often not feasible in studies with vulnerable populations where the number of trials is limited and artifacts are frequent. This necessitates the use of advanced motion artifact correction (MAC) algorithms to preserve statistical power and data integrity [68].
Comparative Efficacy of Motion Correction Techniques
A seminal study directly compared the performance of five motion correction techniques on real fNIRS data containing task-related, low-frequency motion artifacts [68]. The results demonstrated that correcting for motion artifacts is always better than rejecting trials. Among the methods tested, wavelet filtering was the most effective, successfully reducing the artifact in 93% of the cases [68].
Table 2: Performance Comparison of Motion Artifact Correction Techniques
| Correction Technique | Underlying Principle | Key Advantages | Reported Efficacy |
|---|---|---|---|
| Wavelet Filtering [68] | Multi-resolution analysis to isolate and remove artifact components in specific wavelet domains. | Highly effective for spike-type and slow-drift artifacts; does not require auxiliary measurements. | Most effective; 93% success rate in artifact reduction [68] |
| Spline Interpolation [68] | Identifies artifact segments and replaces them with interpolated spline curves based on clean data segments. | Effective for large, isolated spikes; relatively simple implementation. | Performance varies with artifact type and density [68] |
| PCA-based Methods [68] | Removes principal components assumed to be dominated by motion artifacts. | Can remove widespread, global artifacts; but may also remove neural signal. | Less effective than wavelet filtering for task-correlated artifacts [68] |
| Kalman Filtering [68] | Adaptive filtering that uses a state-space model to predict and clean the signal in real-time. | Suitable for online processing; can incorporate a model of the hemodynamic response. | Lower performance compared to wavelet and spline methods [68] |
| CBSI [68] | Utilizes the negative correlation between HbO and HbR during brain activation to correct artifacts. | Simple and computationally efficient; assumes a fixed hemodynamic relationship. | Less effective for complex or large-amplitude artifacts [68] |
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Materials and Methods for Artifact Management in fNIRS Research
| Item / Method | Function / Purpose | Technical Specification / Protocol |
|---|---|---|
| Short-Separation Channels [67] | Measures extracerebral hemodynamics from scalp; used as a nuisance regressor to clean standard fNIRS channels. | Source-detector distance of ~0.8 cm for adults; should be placed near standard long-separation channels [67]. |
| Physiological Monitoring (SPA-fNIRS) [64] | Records systemic physiological confounds (e.g., HR, MAP, EtCO₂) for subsequent regression. | Requires integrated acquisition of: • Pulse Oximeter (Heart Rate) • Finapres/BioPac (Mean Arterial Pressure) • Capnograph (End-Tidal CO₂) [64] [67]. |
| Wavelet Filtering Algorithm [68] | Corrects motion artifacts by decomposing the signal and thresholding wavelet coefficients associated with artifacts. | Implemented in toolboxes like Homer2 and NIRS Brain AnalyzIR; optimal for low-frequency, task-correlated motion [68]. |
| Recursive Least-Squares Estimator (RLSE) [66] | Adaptive filter for online/offline removal of physiological and superficial noises. | Models signal as: Expected HR + SSS data + Physiological noises (sine/cosine) + Baseline drift [66]. |
| Where's Wally Paradigm [8] | A standardized neurovascular coupling (NVC) test to evoke a controlled hemodynamic response in the prefrontal cortex. | Participant searches for "Wally" in complex scenes: • 5 cycles of 20s eyes-closed / 40s eyes-open • fNIRS placed 1cm above eyebrow [8]. |
Advanced Modeling and Reproducibility Considerations
Computational modeling provides critical insights into how systemic confounds can lead to deceptive results. An extended computational model of cerebral physiology, "BrainSignals," has demonstrated that changes in blood pressure and CO₂ can produce hemodynamic responses in fNIRS that are virtually indistinguishable from true functional activation [65]. This reinforces the necessity of recording these parameters during experiments.
Furthermore, the reproducibility of fNIRS findings is highly dependent on data quality and analytical choices. The FRESH initiative, a large-scale reproducibility study, found that while nearly 80% of research teams agreed on group-level results for strong hypotheses, agreement at the individual level was lower and improved significantly with better data quality [69]. The primary sources of variability were the handling of poor-quality data, response modeling, and statistical analysis, underscoring the need for clearer methodological and reporting standards in the field [69].
Figure 2: Conceptual diagram of neurovascular coupling and its confounding by systemic physiology, showing how both neural and non-neural sources contribute to the final measured signal.
Accurate interpretation of fNIRS and fMRI data within the framework of neurovascular coupling demands a rigorous and systematic approach to managing artifacts. Physiological noise, motion, and systemic confounds are not mere nuisances but significant sources of signal contamination that can fundamentally alter research conclusions and clinical interpretations. The integration of mitigation strategies—such as short-separation channels, SPA-fNIRS, advanced motion correction algorithms like wavelet filtering, and computational modeling—is paramount for enhancing the validity and reproducibility of neuroimaging findings. As the field progresses toward more naturalistic study designs and clinical applications, the development and standardization of these artifact-handling protocols will be crucial for unlocking the full potential of hemodynamic-based functional neuroimaging.
Functional near-infrared spectroscopy (fNIRS) has emerged as a pivotal neuroimaging technique that leverages near-infrared light to measure cortical hemodynamic responses associated with brain activity. This non-invasive technology capitalizes on the optical properties of biological tissues, specifically the differential absorption characteristics of oxygenated (HbO) and deoxygenated hemoglobin (HbR) in the 700-900 nm range—often termed the "optical window"—where light penetration is optimal due to relatively low absorption by water and other chromophores [70]. The fundamental relationship between neural activity and subsequent hemodynamic changes, known as neurovascular coupling (NVC), forms the physiological basis for fNIRS measurements, mirroring principles utilized in functional magnetic resonance imaging (fMRI) [7] [70].
Despite its advantages in portability, cost-effectiveness, and tolerance to motion artifacts, fNIRS confronts a significant challenge: achieving an optimal signal-to-noise ratio (SNR). The SNR directly impacts the reliability of extracting neural signals from contaminating noise sources, including physiological fluctuations (e.g., cardiac pulsation, respiration), motion artifacts, and instrumental noise [71] [72]. This technical guide comprehensively addresses SNR enhancement through synergistic improvements in sensor configuration and advanced signal processing methodologies, contextualized within the framework of neurovascular coupling research critical for neuroscientists and drug development professionals.
Sensor Configuration for Optimal SNR
The physical design and configuration of the fNIRS sensor constitute the first line of defense against noise, establishing the upper limit of signal quality for subsequent processing.
Hardware Design Parameters
Strategic selection of hardware components directly influences the quality of the raw acquired signal. Key design considerations include:
- Source-Detector Separation: A distance of 3 cm is widely adopted for adult studies as it represents an optimal balance, allowing light to penetrate sufficiently to reach the cerebral cortex (typically 1.5-2 cm below the scalp) while maintaining adequate signal intensity [70] [72]. Shorter distances primarily sample superficial tissues and scalp hemodynamics, while longer distances drastically attenuate the light, degrading SNR.
- Wavelength Selection: The choice of specific wavelengths is critical for effectively discriminating between HbO and HbR based on their distinct absorption spectra. Systems commonly employ pairs such as 660/850 nm [72] or 730/850 nm [73], situated within the optical window where the absorption contrast between the two chromophores is maximized.
- Modular and Multichannel Architectures: Distributed multichannel systems enhance spatial sampling and regional accuracy. Recent designs integrate multiple light sources and detectors, forming a dense measurement grid over regions of interest like the prefrontal cortex (PFC). This approach facilitates the capture of comprehensive brain activity patterns and improves resilience to localized signal dropouts [72].
Table 1: Key Hardware Design Parameters for fNIRS SNR Optimization
| Parameter | Typical Configuration | Impact on SNR | Rationale |
|---|---|---|---|
| Source-Detector Distance | 3.0 cm (Adults) | Establishes signal strength and cortical penetration | Balances deep penetration against photon attenuation [70] |
| Wavelengths | 660 nm & 850 nm; 730 nm & 850 nm | Enables chromophore differentiation | Targets peak absorption differences between HbO and HbR [73] [72] |
| System Architecture | Distributed multichannel | Improves spatial resolution and regional accuracy | Captures broader and more robust cortical activity patterns [72] |
Wearable System Innovations
The evolution towards wearable, wireless fNIRS platforms introduces specific SNR challenges, primarily related to motion artifacts and environmental interference. Recent advancements address these issues through:
- Integrated Circuit Design: Utilizing specialized chips like the MAX86141, which integrates control for multiple light sources and detectors with analog-to-digital conversion (ADC). This miniaturization reduces the distance between the subject and the primary signal digitization point, mitigating analog signal degradation [72].
- On-Board Preprocessing: Incorporating Microcontroller Units (MCUs) for preliminary data processing tasks, such as digital signal reordering and temporal alignment, immediately after acquisition. This allows for initial data integrity checks before wireless transmission [72].
- Robust Physical Design: Ensuring stable and consistent optode-scalp contact is paramount. Systems are being designed with ergonomic, form-fitting helmets or headbands to minimize slippage, a major source of motion-induced noise [71].
Signal Processing Techniques for SNR Enhancement
Post-acquisition signal processing is indispensable for isolating the neural signal from the confounding noise inherent in fNIRS data.
Motion Artifact Correction and Filtering
The initial processing stage focuses on removing non-neural physiological noise and motion artifacts.
- Artifact Identification: The Coefficient of Variation (CV) method is a common starting point, where the standard deviation of a channel's raw signal is divided by its mean. Channels with a CV exceeding a set threshold (e.g., 20%) are flagged as noisy, often due to poor contact or significant motion artifacts, and can be excluded or heavily weighted in subsequent processing [73].
- Temporal Filtering: Band-pass filtering is a fundamental step. A high-pass filter (e.g., cut-off around 0.01 Hz) removes very low-frequency baseline drift, while a low-pass filter (e.g., cut-off around 0.1-0.3 Hz) attenuates high-frequency noise like cardiac pulsation (~1 Hz), isolating the slower hemodynamic response [74].
Advanced Computational and AI-Driven Methods
Beyond traditional filtering, advanced computational frameworks leverage the power of machine learning and deep learning to achieve superior SNR enhancement and task decoding.
- Novel Neural Network Architectures: The MetaNIRS framework exemplifies modern approaches. It integrates a long-range dilation multilayer perceptron (LongDilMLP) with a PoolFormer backbone. The LongDilMLP is specifically designed to capture the inherent delay and temporal dependencies of the hemodynamic response using dilated convolutions, which model long-range interactions without a proportional increase in parameters. The PoolFormer replaces the computationally expensive self-attention mechanism of traditional transformers with a simpler, parameter-free pooling operator, making it highly efficient and robust for fNIRS's characteristic smooth signals [75].
- Optimization Frameworks for Wearable Platforms: To address the computational constraints of mobile systems, novel optimization frameworks have been developed. These can reduce the memory footprint of neural network processing by up to 24% while maintaining or improving signal clarity, making high-fidelity, real-time processing feasible on wearable devices [71].
- Signal Variability as a Metric: Brain signal variability, calculated from moment-to-moment fluctuations in HbO concentration, is itself a meaningful biomarker of neural processing efficiency. Processing pipelines are being refined to accurately preserve this variability, as it has been shown to correlate with cognitive load and performance, and is diminished in clinical populations such as older adults with hearing loss [74].
Table 2: Advanced Signal Processing Methods for fNIRS SNR Enhancement
| Method | Core Mechanism | Application Context | Reported Benefit |
|---|---|---|---|
| MetaNIRS Framework [75] | LongDilMLP captures hemodynamic delays; PoolFormer provides efficient token mixing. | Motor Execution/Imagery Decoding | Achieved SOTA classification accuracy (e.g., 84.14% on a self-collected MI dataset). |
| Memory-Optimized NN Framework [71] | Neural network model compression and optimization. | Wearable fNIRS Platforms | 24% reduction in memory footprint while enhancing reconstruction accuracy. |
| Brain Signal Variability Analysis [74] | Analysis of moment-to-moment HbO fluctuations. | Assessing neural resource allocation under cognitive load (e.g., auditory tasks). | Serves as a sensitive biomarker for cognitive state and neural health. |
Experimental Protocols for Validation
Rigorous experimental paradigms are required to validate the efficacy of any SNR enhancement technique. Below is a detailed methodology for a mental arithmetic (MA) task, a common paradigm for probing prefrontal cortex function.
Mental Arithmetic (MA) Paradigm Protocol
Objective: To elicit robust, measurable prefrontal cortex (PFC) activation for system calibration and SNR assessment [72].
Participant Preparation:
- Recruit participants with no history of neurological disorders.
- Position the fNIRS headset ensuring optodes cover the PFC region according to the international 10-20 system. For a multichannel system, use a configuration with at least two light sources (e.g., 680 nm and 850 nm) and one detector per hemisphere, with a fixed source-detector distance (e.g., ~1.8 cm for a compact design) [72].
- Instruct participants to remain still and minimize gross motor movements during the recording.
Experimental Procedure (Block Design):
- Baseline/Rest Block (60 seconds): Participants fixate on a cross-hair and relax their mind without engaging in any structured thought.
- Stimulation/Task Block (30 seconds): Participants are presented with a series of arithmetic problems (e.g., "57 - 14" or "23 * 3") and are instructed to silently calculate the answer as quickly and accurately as possible.
- Repeat this sequence for a minimum of 5-10 cycles to ensure adequate data for averaging and statistical power.
Data Analysis Workflow:
- Preprocessing: Apply a band-pass filter (e.g., 0.01 - 0.2 Hz) to raw intensity data to remove drift and physiological noise.
- Conversion: Transform preprocessed light intensity signals into HbO and HbR concentration changes using the Modified Beer-Lambert Law (MBLL) [70] [72].
- General Linear Model (GLM): Model the hemodynamic response for each channel using a canonical hemodynamic response function convolved with the task block regressor.
- SNR & Activation Assessment: Calculate the contrast between task and baseline blocks. Channels or regions showing a statistically significant increase in HbO during the task period relative to baseline are considered active. The magnitude and consistency of this response across trials serve as a direct metric of system SNR.
The Scientist's Toolkit: Essential Research Reagents & Materials
Successful fNIRS research relies on a suite of specialized tools and analytical solutions. The following table details key components of a modern fNIRS research toolkit.
Table 3: Essential Research Reagents and Solutions for fNIRS Studies
| Tool/Reagent | Function/Description | Example/Specification |
|---|---|---|
| Multichannel fNIRS System | Records hemodynamic signals from multiple brain regions simultaneously. | NirSmart-6000A; Custom systems with 24+ sources/detectors [73] [72]. |
| MAX86141 Integrated Chip | Controls LEDs and detectors, performs ADC. Foundational for compact systems. | Enables integration of 2+ wavelengths (e.g., 680/850 nm) and a detector on a single chip [72]. |
| Prefrontal Cortex (PFC) Probe Set | Specific optode placement to target the prefrontal cortex. | Custom holder or headcap aligning sources & detectors over Fp1, Fpz, Fp2 (10-20 system) [74] [72]. |
| MetaNIRS Software Framework | Deep learning framework for decoding tasks like motor imagery/execution. | Based on PoolFormer and LongDilMLP; available code for classification [75]. |
| Homer2 / NIRS-KIT Toolbox | Open-source software for fNIRS data preprocessing and statistical analysis. | Implemented in MATLAB; standard for filtering, MBLL conversion, and GLM [73]. |
| Mental Arithmetic Stimulus Set | Standardized cognitive tasks to evoke robust PFC activation for system validation. | Series of arithmetic problems (e.g., two-digit subtraction/multiplication) [72]. |
| Coma Recovery Scale-Revised (CRS-R) | Behavioral assessment scale for clinical validation in disorders of consciousness. | Used to correlate fNIRS functional connectivity measures with clinical scores [73] [70]. |
Enhancing the signal-to-noise ratio in fNIRS is not a single-step solution but a comprehensive strategy spanning hardware engineering, sensor design, and sophisticated computational analysis. Optimizing sensor configuration—through precise source-detector geometry, wavelength selection, and robust wearable design—provides the high-fidelity raw data essential for all subsequent stages. Advanced signal processing techniques, particularly those leveraging modern AI frameworks like MetaNIRS and variability analysis, then unlock the full potential of this data by effectively isolating the neural signal, thereby improving the accuracy of brain state decoding.
The continued refinement of these methods is crucial for advancing fundamental research into neurovascular coupling and for broadening the clinical applicability of fNIRS. From diagnosing disorders of consciousness and monitoring cognitive decline to enabling real-time brain-computer interfaces for neurorehabilitation, a high SNR is the linchpin of reliable and interpretable fNIRS data. As the field progresses, the integration of multimodal imaging, standardized processing pipelines, and AI-driven analytics promises to further solidify fNIRS's role as an indispensable tool in neuroscience and pharmaceutical development.
The hemodynamic inverse problem represents one of the most significant challenges in modern cognitive neuroscience. This problem concerns the difficulty of making valid and precise estimates of underlying neural activity from measured hemodynamic responses, which are temporally sluggish and indirect correlates of neuronal events [76]. In functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS), researchers observe vascular changes—specifically, fluctuations in cerebral blood flow, volume, and oxygenation—that occur seconds after neuronal firing and last for tens of seconds [76]. The central challenge lies in working backward from these observed vascular phenomena to infer the timing, amplitude, and spatial location of the neural activity that generated them. This technical guide examines the current state of inverse problem methodologies within the framework of neurovascular coupling (NVC) research, providing researchers and drug development professionals with advanced tools for interpreting hemodynamic signals in both basic and clinical contexts.
Fundamental Concepts
Neurovascular coupling (NVC) describes the biological mechanism that links neural activity to subsequent changes in local cerebral blood flow [11]. This process ensures that active brain regions receive adequate oxygen and nutrients to support their metabolic demands. The neurovascular unit (NVU)—a functional complex comprising neurons, astrocytes, vascular smooth muscle cells, and pericytes—orchestrates this precise coordination [11]. When neurons become active, they trigger vasodilation either directly by stimulating vascular cells or indirectly through astrocytic signaling, leading to a rapid increase in local blood flow that forms the basis for both fMRI and fNIRS measurements [11].
The hemodynamic inverse problem arises from the fundamental disconnect between the rapid timescale of neural events (milliseconds) and the slow temporal dynamics of the hemodynamic response (seconds) [76]. While cognitive acts can be completed in under a second, the accompanying hemodynamic response begins seconds after the neuronal event and lasts for tens of seconds [76]. This temporal mismatch, combined with the complex nonlinear transformations between neural activity and vascular responses, creates a challenging inference problem for researchers attempting to deduce precise neural timing and amplitude from observed hemodynamic signals.
Clinical and Research Significance
Understanding and addressing the inverse problem has profound implications for both basic neuroscience and drug development. NVC dysfunction has been implicated in various neurological conditions, including Alzheimer's disease [7], Parkinson's disease [77], stroke, and cerebral small vessel disease [11]. In Parkinson's disease, for example, researchers have identified NVC alterations in the striato-thalamo-cortical motor circuit, and have observed that levodopa treatment primarily restores normal NVC by modulating neuronal activity rather than directly affecting blood flow [77]. For drug development professionals, accurate solutions to the inverse problem enable more precise assessment of how experimental therapeutics affect neural circuitry and neurovascular function.
Methodological Approaches to the Inverse Problem
Modeling the Hemodynamic Response Function
The hemodynamic response function (HRF) models the relationship between neural activity and the observed blood oxygenation level-dependent (BOLD) signal in fMRI. Most analytical approaches assume that the BOLD response can be modeled as a linear time-invariant system, where the signal at time (t), (y(t)), is represented as the convolution of a stimulus function (s(t)) and the hemodynamic response (h(t)):
[ y(t) = (s \ast h)(t) ]
In practice, (h(t)) is frequently modeled as a linear combination of basis functions (g_i(t)):
[ h(t) = \sum{i=1}^B \betai g_i(t) ]
where (\beta_i) are unknown parameters estimated from the data [78].
Table 1: Comparison of Hemodynamic Response Modeling Approaches
| Model Type | Key Features | Advantages | Limitations |
|---|---|---|---|
| Canonical HRF | Single predetermined shape | Simple, minimal parameters | Inflexible, poor fit to variant responses |
| Basis Set Functions | Linear combination of smooth functions (e.g., gamma functions) | Flexible, accommodates response variability | Potential parameter confusability |
| Finite Impulse Response (FIR) | Separate parameter for each time point post-stimulus | Maximum flexibility | Low statistical power, many parameters |
| Nonlinear Estimation | Direct parameter estimation of height, delay, dispersion | Direct parameters of interest | Computationally intensive |
Parameter Estimation and Inference
When analyzing the shape of the estimated HRF, researchers typically extract summary measures of psychological interest, including:
- Amplitude/Height (H): Related to the intensity of neural activity
- Time-to-Peak (T): Provides information about onset latency of activation
- Full-Width at Half-Max (W): Offers insights into the duration of neural activity [78]
Accurate estimation of these parameters is complicated by the fact that the BOLD response behaves as a nonlinear integrator, as the vascular response saturates over time [78]. This nonlinearity means that changes in the duration of neuronal activity can affect both the amplitude and duration of the evoked BOLD response, creating challenges for straightforward interpretation.
Advanced Mathematical Frameworks
For fNIRS data, the inverse problem involves reconstructing internal optical properties from boundary measurements. The fundamental equation governing light propagation in biological tissue is the radiative transport equation (RTE):
[ \frac{\partial}{c\partial t}I(r,\Omega,t) + \Omega \cdot \nabla I(r,\Omega,t) + \mua(r) + \mus(r)I(r,\Omega,t) = \mus(r)\int{4\pi}d\Omega'P(r,\Omega,\Omega')I(r,\Omega',t) + q(r,\Omega,t) ]
where (I(r,\Omega,t)) represents energy radiance, (\mua(r)) and (\mus(r)) are absorption and scattering coefficients, and (P(r,\Omega,\Omega')) is the scattering phase function [79].
In practice, this complex equation is often simplified using the diffusion approximation, which assumes that radiance in optically thick media with multiple scattering is nearly isotropic. This approximation leads to the diffusion equation:
[ \frac{\partial}{c\partial t}\Phi(r,t) - \nabla \cdot \kappa(r)\nabla \Phi(r,t) + \mu_a(r)\Phi(r,t) = q(r,t) ]
where (\Phi(r,t)) denotes the fluence rate and (\kappa(r)) is the diffusion coefficient [79].
The inverse problem in fNIRS can then be linearized as:
[ y = Ax ]
where (A) represents the sensitivity matrix, (y) is the difference in optical density measurements, and (x) denotes the change in absorption coefficient [79].
Experimental Protocols for Investigating the Inverse Problem
Multimodal Integration Approaches
Integrating multiple neuroimaging modalities provides a powerful approach to addressing the inverse problem by leveraging the complementary strengths of different techniques. The fusion of fNIRS and fMRI data has emerged as a particularly valuable methodology for revealing spatiotemporal dynamics of hemodynamic responses with high resolution across both space and time [80].
Joint Independent Component Analysis (jICA) has been successfully adapted to identify linked components between fNIRS and fMRI datasets. This approach assumes that joint spatial and temporal independences of fNIRS and fMRI satisfy the following generative model:
[ X{fNIRS} = AS{fNIRS},\quad X{fMRI} = AS{fMRI} ]
where (X{fNIRS}) and (X{fMRI}) represent the group data from fNIRS and fMRI respectively, (S{fNIRS}) and (S{fMRI}) are the sources, and (A) is a shared mixing matrix [80].
The joint decomposition can be rewritten as a single matrix equation:
[ \begin{bmatrix} X{1fNIRS} & X{1fMRI} \ X{2fNIRS} & X{2fMRI} \ \vdots & \vdots \ X{nfNIRS} & X{nfMRI} \end{bmatrix} = A \begin{bmatrix} S{1fNIRS} & S{1fMRI} \ S{2fNIRS} & S{2fMRI} \ \vdots & \vdots \ S{nfNIRS} & S{nfMRI} \end{bmatrix} ]
The infomax ICA algorithm is then employed to calculate the shared unmixing matrix (W) (the inverse of (A)) using the following update equation:
[ \Delta W = \eta{I - 2y{fNIRS}(u{fNIRS})^T - 2y{fMRI}(u{fMRI})^T}W ]
where (y{fNIRS} = g(u{fNIRS})), (y{fMRI} = g(u{fMRI})), (u{fNIRS} = WX{fNIRS}), (u{fMRI} = WX{fMRI}), and (g(x) = 1/(1+e^{-x})) is the nonlinearity in the neural network [80].
Protocol: fNIRS-fMRI Fusion in Motor Tasks
A representative experimental protocol for fNIRS-fMRI fusion involves right finger tapping tasks with the following parameters:
fNIRS Acquisition:
- System: 24-channel fNIRS system (Oxymon MKIII, Artinis) with 8 sources and 8 detectors
- Wavelengths: 781 nm and 856 nm
- Design: Block design with 21s activation alternated with 30s rest, repeated 10 times
- Total Recording Time: 552 seconds
- Sampling Rate: 9.75 Hz
- Data Processing: Conversion to ΔHbO₂ and ΔHbR concentrations using differential pathlength factor (DPF=4) [80]
fMRI Acquisition:
- System: 3.0 T MRI scanner
- Sequence: Echo planar imaging (EPI)
- Parameters: TR/TE = 3000/35 ms, flip angle = 80°, 35 slices, 4 mm slice thickness [80]
Data Analysis Pipeline:
- Segmentation: Extract epochs from -35s prior to event onset until 35s after event code
- Channel Selection: Identify channels near left motor cortex (channels 8-12)
- Signal Averaging: Combine data from selected channels to improve signal-to-noise ratio
- Component Analysis: Apply jICA to identify linked fNIRS and fMRI components
- Spatiotemporal Visualization: Generate "snapshots" and movies of hemodynamic dynamics
Table 2: Research Reagent Solutions for Neurovascular Coupling Studies
| Reagent/Resource | Function | Application Context |
|---|---|---|
| Oxymon MKIII fNIRS System | Measures HbO₂ and HbR concentration changes | Multimodal imaging protocols |
| 3.0 T MRI Scanner | Acquires BOLD signal with high spatial resolution | Multimodal imaging protocols |
| SPM Software | Statistical parametric mapping for fMRI analysis | Data processing and analysis |
| CSCKF-CSCKS Algorithm | Confounds Square-root Cubature Kalman Filtering/Smoothing | Hemodynamic model parameter estimation |
| Joint ICA Toolbox | Independent component analysis for multimodal data | fNIRS-fMRI data fusion |
Cellular-Level Investigation Protocols
Understanding the inverse problem requires knowledge of the cellular mechanisms underlying neurovascular coupling. Key experimental approaches include:
Optogenetic Stimulation in Awake Mice:
- Preparation: Cranial window implantation for optical access
- Stimulation: Cell-type-specific optogenetic activation of neurons or astrocytes
- Imaging: Two-photon microscopy to measure vascular diameter and blood flow changes
- Genetic Manipulation: Cell-specific knockout of signaling molecules (e.g., Cx37, Cx40 in endothelial cells) [81]
Pharmacological Dissection of Signaling Pathways:
- Receptor Blockers: Application of glutamatergic, GABAergic, cholinergic, or nitrergic antagonists
- Enzyme Inhibitors: Inhibition of nitric oxide synthase, cyclooxygenase, or cytochrome P450 epoxygenase
- Measurement: Combined assessment of neural activity (LFP, MUA) and hemodynamic responses [11]
Signaling Pathways in Neurovascular Coupling
The cellular mechanisms of neurovascular coupling involve complex signaling between neurons, astrocytes, and vascular cells. Understanding these pathways is essential for developing accurate solutions to the inverse problem.
Neuronal Signaling Components:
- Glutamatergic Neurons: Pyramidal neurons serve as "neurogenic hubs" in NVC, releasing glutamate that activates both astrocytes and interneurons [11]
- GABAergic Interneurons: Modulate the output of excitatory pyramidal cells, primarily through indirect regulation of hemodynamic responses [11]
- Nitric Oxide Synthase (NOS) Expressing Neurons: Neurons expressing neuronal NOS (nNOS) significantly influence both resting and reactive arterial dimensions through nitric oxide release [11]
Astrocytic Signaling:
- Calcium Signaling: Activation of metabotropic glutamate receptors (mGluR5) triggers Ca²⁺ release from internal stores
- Vasoactive Factor Release: Calcium waves lead to production of vasoactive substances including prostaglandins (PGE₂) and epoxyeicosatrienoic acids (EETs) [11]
Vascular Response Propagation:
- Endothelial Gap Junctions: Composed of connexins (Cx37, Cx40) that enable long-range propagation of vasodilation signals through the vasculature [81]
- Arterio-venous Zonation: Connexin expression and cell-cell coupling strength vary along the arterio-venous axis, with arteries being the most strongly coupled segment [81]
Clinical Applications and Therapeutic Implications
Accurate solutions to the hemodynamic inverse problem have significant clinical applications, particularly in drug development for neurological disorders. Quantitative assessment of neurovascular coupling provides valuable biomarkers for early diagnosis, treatment monitoring, and therapeutic development.
NVC Alterations in Neurological Disorders
Parkinson's Disease:
- Affected Regions: NVC alterations occur in the striato-thalamo-cortical motor circuit and motor control regions
- Neurotransmitter Systems: Dysfunction involves dopaminergic, serotoninergic, opioid, and cholinergic systems
- Levodopa Effects: Levodopa ameliorates abnormal NVC primarily by modulating neuronal activity (ALFF changes) with limited effects on blood perfusion (CBF modifications) [77]
Alzheimer's Disease:
- Mechanism: Amyloid-β deposition adversely affects endothelial function and pericyte signaling
- Consequence: Compromises the NVU's ability to match blood flow to neural demand [7]
Cerebrovascular Diseases:
- Stroke and SAH: Disruption of neurovascular coupling contributes to brain dysfunction and neuronal damage
- Therapeutic Target: Functional reconstruction of NVC promotes cell survival, debris removal, and microcirculation reconstruction [11]
Advanced Algorithmic Approaches
Recent advancements in numerical methods have improved our ability to solve the inverse problem:
Hemodynamic Model Solving Algorithms:
- Approach: Confounds Square-root Cubature Kalman Filtering and Smoothing (CSCKF-CSCKS)
- Reported Accuracy: 84% in estimating biophysical parameters and hidden states
- Improvement Strategy: Increasing maximum iterations can improve accuracy by 5.76% [82]
Inverse Problem Solutions in fNIRS:
- Methods: Algebraic reconstruction technique, model-based reconstruction, singular value decomposition, Tikhonov regularization, and Bayesian methods
- Challenge: Ill-posed nature requires regularization techniques for stable solutions [79]
The field of inverse problem solving in neurovascular research continues to evolve with several promising directions:
Integration of Artificial Intelligence: Machine learning and deep learning approaches are being increasingly applied to solve the inverse problem, potentially offering more accurate mappings between hemodynamic signals and neural activity without relying on simplified analytical models.
Multi-Omics Integration: Combining transcriptomic, proteomic, and metabolomic data with hemodynamic measurements may provide new insights into the molecular mechanisms underlying neurovascular coupling and its alterations in disease states.
High-Resolution Imaging: Advances in imaging technology, including ultra-high field MRI and super-resolution microscopy, will provide more detailed data on neurovascular interactions at cellular and subcellular levels.
Personalized Hemodynamic Modeling: Development of subject-specific biophysical models that incorporate individual anatomical, physiological, and genetic factors may improve the accuracy of inverse problem solutions in both research and clinical applications.
In conclusion, addressing the inverse problem from hemodynamic signals to neural activity remains a challenging but essential endeavor in cognitive neuroscience and neuropharmacology. The continued development of sophisticated mathematical models, multimodal integration approaches, and cellular-level mechanistic studies will enhance our ability to accurately infer neural activity from non-invasive hemodynamic measurements, ultimately advancing both basic understanding of brain function and clinical applications in neurological and psychiatric disorders.
Neurovascular coupling (NVC), the fundamental process that links neuronal activity to subsequent changes in cerebral blood flow, has emerged as a critical biomarker in neuroscience and drug development research [23]. The accurate assessment of NVC using functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS) relies heavily on the precise interplay between task design and baseline physiological stability. However, the proliferation of divergent experimental methodologies across research groups has created significant challenges in interpreting and comparing NVC findings, particularly due to variations in task complexity and inadequate control of baseline conditions [83]. Within the broader context of neurovascular coupling research, this technical guide addresses the fundamental need for standardized protocols that can yield robust, reproducible NVC measurements essential for both basic science and clinical applications, including the evaluation of pharmacological interventions.
The integrity of NVC assessment rests on two pillars: first, the selection of activation tasks that selectively engage target cortical regions without introducing confounding neural activation; and second, the maintenance of stable baseline physiological conditions that ensure any measured hemodynamic changes reflect true neurovascular function rather than systemic artifacts [23]. This guide synthesizes current evidence to provide researchers with optimized methodologies for both task design and baseline stability, supported by quantitative data and practical experimental workflows.
Neurovascular Coupling Mechanisms and Assessment Principles
The Neurovascular Unit and Functional Hyperemia
The neurovascular unit forms the anatomical and functional basis of NVC, comprising three principal components: vascular smooth muscle cells, neurons, and astrocyte glial cells [23]. This triad operates in concert to ensure precise regional cerebral blood flow (CBF) regulation in response to neuronal metabolic demands. Upon neuronal activation, glutamate release triggers both direct vasoactive signaling from neurons and indirect pathways through astrocytes. Neurons activate neuronal nitric oxide synthase (nNOS), producing nitric oxide (NO) that directly dilates parenchymal arterioles, while astrocytes generate arachidonic acid metabolites including vasodilatory epoxyeicosatrienoic acids (EETs) and prostaglandins, alongside vasoconstrictive compounds [23].
This coordinated signaling results in functional hyperemia, characterized by a region-specific increase in CBF that delivers oxygen and glucose to active neurons while removing metabolic byproducts [23]. Under normal conditions, this neurovascular response demonstrates a 4-fold greater increase in CBF relative to the increase in ATP consumption, creating the theoretical foundation for both blood-oxygen-level-dependent (BOLD) fMRI and fNIRS signal generation [23].
Assessment Techniques: fMRI and fNIRS
fMRI measures the BOLD signal, which reflects the balance between cerebral blood flow, blood volume, and oxygen metabolism. The technique leverages the magnetic properties of hemoglobin, with deoxygenated hemoglobin acting as an endogenous contrast agent due to its paramagnetic properties [23].
fNIRS employs near-infrared light (700-1000 nm) to measure concentration changes in oxygenated (Δ[HbO]) and deoxygenated hemoglobin (Δ[HbR]) in the cortical microvasculature [84]. During neuronal activation, neurovascular coupling triggers increased arterial blood flow to the active region, resulting in an increase in HbO and a decrease in HbR concentrations [84]. The hemodynamic response function (HRF) measured by fNIRS typically shows a 2-second delay after neural activation onset, reaching a plateau approximately 6-10 seconds post-stimulus [32] [85].
Table 1: Comparison of NVC Assessment Modalities
| Parameter | fMRI | fNIRS |
|---|---|---|
| Primary Signal | BOLD (Blood Oxygen Level Dependent) | HbO/HbR concentration changes |
| Spatial Resolution | High (millimeter range) | Moderate (limited to cortical surfaces) |
| Temporal Resolution | Moderate (seconds) | High (up to 10 Hz) |
| Portability | Low (fixed systems) | High (portable/wireless systems) |
| Environment | Restrictive | Flexible (tolerates more movement) |
| Depth Penetration | Whole brain | Superficial cortex (2-3 cm) |
Task Design Optimization for Selective Cortical Activation
Visual Task Complexity and Specificity
Visual stimulation paradigms remain the most common approach for NVC assessment, particularly when targeting the posterior cerebral artery (PCA) territory supplying the visual cortex. However, significant differences in task complexity dramatically influence the selectivity and magnitude of NVC responses [83].
Simple versus Complex Visual Tasks: Evidence indicates that simple visual tasks (e.g., passive viewing of a moving target) activate the posterior circulation most selectively, while complex visual tasks (e.g., pattern recognition, active visual search) robustly activate both posterior and middle cerebral artery (MCA) territories, thereby reducing selectivity for the visual cortex [83]. This reduced selectivity occurs because complex tasks engage additional brain regions beyond the primary visual cortex, including parietal areas involved in visual processing and attention, which are partially perfused by the PCA [83].
Eye Movement Patterns: The amplitude and speed of eye movements during visual tasks significantly impact NVC measurements. Studies demonstrate that greater visual task amplitude (larger eye movements) increases selectivity for the posterior circulation, while varying visual target speed has minimal effect on posterior circulation activation [83].
Table 2: Impact of Visual Task Parameters on NVC Selectivity
| Task Parameter | Effect on PCA Selectivity | Effect on MCA Activation | Recommendation |
|---|---|---|---|
| Task Complexity | Simple tasks increase selectivity | Complex tasks increase non-specific MCA activation | Use simple passive viewing tasks |
| Eye Movement Amplitude | Larger amplitudes increase selectivity | Minimal effect | Incorporate larger visual targets requiring >5° eye movements |
| Eye Movement Speed | No significant effect | No significant effect | Speed can be adjusted based on participant comfort |
| Cognitive Demand | Low demand increases selectivity | High demand increases frontal lobe activation | Avoid pattern recognition, counting, or decision-making tasks |
Alternative Sensory Modalities: Auditory Stimulation Paradigms
For studies targeting non-visual cortical areas, auditory stimulation provides a valuable alternative. Intensity-dependent amplitude changes (IDAP) in auditory paradigms have demonstrated robust NVC responses measurable with both EEG and fNIRS [32] [85].
Research shows that increasing auditory intensity (e.g., from 70.9 dB to 94.5 dB) produces corresponding increases in N1 and P2 event-related potential components alongside increased HbO and decreased HbR concentrations in auditory and prefrontal cortices [32]. However, extremely high intensities (≥94.5 dB) may induce paradoxical decreases in HbO, potentially due to systemic vasoconstriction [32] [85]. The relationship between sound intensity and hemodynamic response appears modulated by perceived loudness rather than physical intensity alone, highlighting the importance of subjective perception in auditory NVC paradigms [85].
Motor and Imagery Tasks
Motor execution and motor imagery tasks engage the supplementary motor area (SMA) and primary motor cortex (M1), providing reliable NVC responses in frontal regions. Studies comparing fNIRS with fMRI have validated that both motor execution and kinesthetic motor imagery produce detectable SMA activation, with motor execution generating stronger responses than imagery alone [86].
For clinical populations with movement limitations, motor imagery offers particular value. During motor imagery, participants mentally simulate movements without physical execution, generating measurable hemodynamic responses in motor regions [87] [86]. Whole-body motor imagery may produce more spatially specific SMA activation compared to hand movement imagery, suggesting task-specific optimization opportunities [86].
Baseline Stability and Physiological Considerations
Controlling for Confounding Factors
Baseline physiological stability is paramount for reliable NVC assessment, as numerous systemic factors can profoundly influence cerebral hemodynamics independent of neural activity:
Arterial Blood Gases: Cerebral blood flow exhibits exquisite sensitivity to arterial carbon dioxide (CO₂) partial pressure, with a 1-mm Hg change from eupneic CO₂ levels altering CBF by 3-6% [23]. Maintaining stable respiration and monitoring end-tidal CO₂ during NVC assessments is crucial, particularly in populations with altered respiratory physiology.
Cardiovascular Influences: Heart rate, blood pressure, and Mayer waves (cyclic changes in arterial blood pressure) introduce substantial noise in fNIRS signals [84]. Baseline cardiovascular monitoring and appropriate signal processing techniques are essential to distinguish true NVC responses from systemic cardiovascular artifacts.
Autonomic Regulation: Both sympathetic and parasympathetic nervous systems influence cerebrovascular tone, though their impact on dynamic cerebral autoregulation appears modest in humans [23]. Standardizing testing conditions to minimize autonomic fluctuations optimizes measurement reliability.
State-Dependent NVC Variability
Emerging evidence indicates that NVC is not static but exhibits state-dependent variability, even in healthy individuals [88]. Studies comparing eye-open-eye-close tasks with resting states demonstrate distinctive patterns of EEG-fMRI spectral correspondence across conditions [88]. During active tasks, phase-amplitude coupling dominates between EEG alpha-band oscillations and low-frequency fMRI signals in visual areas, while resting states show amplitude-amplitude coupling between EEG gamma-band and high-frequency fMRI signals [88]. These findings underscore the importance of standardizing participant state and behavioral context during NVC assessments.
Experimental Protocols and Methodological Guidelines
Optimized Visual Task Protocol
Based on current evidence, the following visual task protocol optimizes selectivity for posterior circulation assessment:
- Stimulus Characteristics: Use a simple moving target (e.g., circle) against a neutral background. Avoid patterns, complex shapes, or decision-making elements.
- Eye Movement Parameters: Implement larger amplitude movements (>5° visual angle) at teleologically relevant speeds (approximately 20-40°/sec).
- Task Structure: Employ block designs with 30-second baseline periods followed by 30-second activation epochs. Ensure adequate rest between blocks (≥60 seconds) to allow hemodynamic recovery.
- Participant Instruction: For passive tasks, instruct participants to simply follow the target without additional cognitive engagement. Avoid counting or pattern identification tasks.
- Baseline Recording: Ensure stable cardiovascular and respiratory measures during baseline, discarding epochs with significant deviation from resting norms.
Integrated fNIRS-EEG Auditory Protocol
For auditory cortex NVC assessment:
- Stimulus Parameters: Utilize pure tones at 77.9 dB, 84.5 dB, and 89.5 dB intensities, presented in randomized order. Stimulus duration of 500 ms with interstimulus intervals of 1-2 seconds.
- Paradigm Structure: Present each intensity 54 times in random order across the recording session. Include silent baseline periods before and after stimulus blocks.
- Simultaneous Recording: Collect continuous fNIRS from auditory and prefrontal cortices alongside EEG recording for N1/P2 component analysis.
- Participant Instruction: Direct participants to passively listen to tones without active response to minimize cognitive confounds.
Data Processing and Quality Control
fNIRS Pre-processing: Implement bandpass filtering (0.01-0.5 Hz) to remove cardiac and respiratory artifacts while preserving the hemodynamic response [84]. Apply wavelet filtering to address motion artifacts while preserving signal integrity. For block designs, incorporate baseline normalization using the pre-stimulus period.
fNIRS Processing: Utilize the general linear model (GLM) for statistical analysis of HbO and HbR responses [84]. For clinical applications, consider incorporating functional connectivity measures such as phase-locking value to assess network integrity [87].
Quality Metrics: Establish pre-defined criteria for signal quality, including signal-to-noise ratio thresholds (>10 dB), head-motion tolerance limits (<5 mm movement), and physiological stability benchmarks (heart rate variation <10%, end-tidal CO₂ variation <3 mm Hg).
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Key Research Reagents and Materials for NVC Studies
| Item | Function/Application | Examples/Specifications |
|---|---|---|
| fNIRS Systems | Continuous-wave systems for measuring HbO/HbR concentration changes | Commercially available systems (e.g., NIRx, Hitachi) with 10+ Hz sampling rate |
| fMRI-Compatible Visual Presentation Systems | Precise stimulus delivery in scanner environment | MR-compatible goggles or projection systems with accurate timing synchronization |
| Auditory Stimulation Equipment | Controlled sound presentation for auditory paradigms | MRI-compatible headphones with calibrated intensity output |
| Physiological Monitoring | Baseline stability assessment and artifact correction | Pulse oximeter, capnometer, blood pressure monitor |
| Analysis Software | Standardized processing of NVC data | iNVC Software (Innovate Calgary), Homer2, NIRS-SPM, AtlasViewer |
| Eye Tracking | Verification of task compliance and eye movement parameters | MRI-compatible eye tracking systems (60+ Hz sampling) |
| Motor Task Apparatus | Standardized motor execution and imagery paradigms | fMRI-compatible response devices, hand dynamometers |
Signaling Pathway and Experimental Workflow
Diagram 1: Neurovascular Coupling Signaling Pathway and Assessment Workflow
Diagram 2: Optimized Experimental Workflow for Robust NVC Assessment
Protocol optimization for robust NVC assessment requires meticulous attention to both task design parameters and baseline physiological stability. The evidence presented demonstrates that simple, well-controlled activation paradigms yield more selective and interpretable NVC responses compared to complex tasks that engage multiple neural networks. Furthermore, maintaining stable baseline conditions through rigorous monitoring and control of cardiovascular and respiratory parameters is essential for distinguishing true neurovascular coupling from systemic confounds.
The standardized approaches outlined in this guide provide a foundation for reproducible NVC assessment across research sites and clinical studies. As NVC continues to gain prominence as a biomarker in drug development and neurological disease management, adherence to these optimized protocols will enhance data quality, improve cross-study comparisons, and accelerate the translation of NVC research into clinical applications. Future methodological developments should focus on further refining task specificity, enhancing signal processing techniques, and establishing consensus standards for NVC assessment across the research community.
Neurovascular coupling (NVC) describes the precise physiological mechanism whereby transient neural activity leads to localized changes in cerebral blood flow (CBF), a process critical for meeting the brain's moment-to-moment metabolic demands [12] [89]. This fundamental process forms the basis for many functional neuroimaging techniques, including functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS), which infer neural activity by measuring hemodynamic responses [90]. Traditionally, NVC has been modeled as a static, invariant relationship between neural activity and hemodynamic response. However, emerging evidence demonstrates that NVC is inherently dynamic and state-dependent, varying significantly based on brain states, physiological conditions, and pathological status [91] [92]. This dynamic nature presents both challenges and opportunities for neuroimaging research and its clinical applications.
The concept of state-dependent NVC changes has profound implications for interpreting neuroimaging data. Assumptions of constant coupling can lead to misinterpretation of brain activity measurements, particularly when comparing different physiological states or patient populations. Recognizing NVC as a dynamic process enables more accurate modeling of neurovascular relationships and enhances the validity of functional brain imaging conclusions [91] [12]. This technical guide explores the mechanisms, assessment methodologies, and functional implications of dynamic NVC, providing researchers with the framework needed to account for state-dependent coupling changes in fMRI and fNIRS research.
Physiological Basis of Dynamic NVC
Cellular and Molecular Mechanisms
At the cellular level, NVC is mediated by the neurovascular unit (NVU), comprising neurons, astrocytes, vascular smooth muscle cells, pericytes, and endothelial cells [89]. This coordinated cellular network regulates cerebral blood flow through intricate signaling pathways. During neuronal activation, glutamate release triggers calcium influx in postsynaptic neurons and adjacent astrocytes, initiating vasoactive signaling cascades [12]. Astrocytes play a particularly crucial role as intermediaries, detecting neuronal activity through their endfeet enveloping cerebral vessels and releasing various vasoactive factors [89].
The dynamic nature of NVC arises from the complex interplay of multiple signaling systems, each with distinct temporal characteristics and contextual dependencies. Key signaling molecules include nitric oxide (NO) synthesized by neuronal and endothelial nitric oxide synthase, prostaglandin E2 (PGE2) released by astrocytes, and calcium ions (Ca²⁺) that directly regulate vascular tone [89]. The relative contribution of each pathway varies across brain regions, physiological states, and pathological conditions, creating the foundation for state-dependent coupling. For instance, research indicates that PGE2 plays a more prominent role in cortical regions, while NO dominates in hippocampal neurovascular coupling [89].
Neural Oscillations and Hemodynamic Coupling
The relationship between specific neural oscillation patterns and hemodynamic responses represents another dimension of NVC dynamics. Different frequency bands correlate distinctly with vascular responses: alpha oscillations (8-12 Hz) typically show negative correlation with the blood oxygen level-dependent (BOLD) signal, while gamma oscillations (30-80 Hz) demonstrate positive correlations [91] [93]. These relationships are not fixed but vary with behavioral states and cognitive demands. During sleep inertia, for example, the coupling between EEG alpha-vigilance markers and thalamic BOLD signals changes progressively as alertness recovers, demonstrating temporal dynamics in neurovascular relationships [91].
Table 1: Key Signaling Molecules in Neurovascular Coupling
| Signaling Molecule | Primary Source | Vascular Effect | Temporal Dynamics |
|---|---|---|---|
| Nitric Oxide (NO) | Neurons, Endothelial cells | Vasodilation | Fast response (seconds) |
| Prostaglandin E2 (PGE2) | Astrocytes | Vasodilation | Intermediate response |
| Calcium ions (Ca²⁺) | Neurons, Astrocytes | Regulates vasoactive release | Rapid, activity-dependent |
| Potassium ions (K⁺) | Astrocytes, Neurons | Vasodilation | Fast spatial buffering |
| Arachidonic acid | Astrocytes | Vasoconstriction | Slow, modulatory role |
Methodological Approaches for Assessing Dynamic NVC
Multimodal Integration Strategies
Comprehensive assessment of dynamic NVC requires integrating complementary neuroimaging modalities that capture both neural activity and hemodynamic responses with appropriate temporal and spatial resolution [92] [90]. The most common approaches combine electrophysiological measures (EEG, MEG) with hemodynamic monitoring (fMRI, fNIRS), leveraging their complementary strengths to characterize coupling dynamics [18] [90].
EEG-fMRI combinations exploit fMRI's high spatial resolution and whole-brain coverage with EEG's millisecond temporal precision, allowing researchers to correlate specific neural oscillatory patterns with BOLD signal changes across distributed brain networks [91] [92]. Meanwhile, EEG-fNIRS integration offers practical advantages for studying naturalistic behaviors and clinical populations, as fNIRS is more tolerant of movement and can be deployed in more ecological settings [32] [8] [18]. For comprehensive vascular assessment, EEG-fNIRS-TCD combinations provide a unique window into multi-level vascular responses, measuring microvascular oxygenation (fNIRS), conduit artery velocity (TCD), and neural activity (EEG) simultaneously [62].
Analytical Frameworks for Dynamic NVC
Advanced analytical methods are essential for quantifying state-dependent NVC changes. Time-lagged correlation analysis identifies optimal temporal relationships between electrophysiological and hemodynamic signals, revealing how coupling latencies vary across states [91] [89]. Task-related component analysis (TRCA) enhances signal-to-noise ratio by extracting reproducible components from bimodal signals, improving characterization of neural patterns underlying NVC [18]. Cross-correlation analysis between hemodynamic signals and systemic physiological measures (blood pressure, end-tidal CO₂) helps disambiguate neurally-driven vascular responses from systemic influences [62].
Machine learning approaches, such as multimodal source power comodulation (mSPoC), facilitate fusion of EEG and fNIRS modalities by extracting components that maximize covariance between electrophysiological and hemodynamic features [93]. These methods enable researchers to identify state-specific coupling patterns that might be obscured in conventional analyses assuming static neurovascular relationships.
Diagram 1: Multimodal Analysis Workflow for Dynamic NVC
Experimental Evidence of State-Dependent NVC Changes
Sleep-Wake Transitions
The sleep-wake cycle provides a compelling paradigm for studying state-dependent NVC. Research using simultaneous EEG-fMRI during sleep inertia (the transitional state of impaired alertness upon awakening) has demonstrated dynamic changes in coupling as alertness recovers [91]. Specifically, the temporal relationship between EEG alpha-vigilance and thalamic BOLD signals evolves progressively across post-awakening sessions, with peak correlation lags shifting as sleep inertia dissipates. Similarly, coupling between the EEG spectral slope (1/f component) and BOLD activity in the anterior cingulate cortex shows time-varying patterns during this transition period [91]. These findings challenge the assumption of static NVC and highlight the need to account for vigilance states when interpreting neurovascular correlations.
Cognitive and Motor Task Demands
Cognitive-motor interference paradigms reveal how NVC dynamics shift under divided attention. During cognitive-motor dual-tasks, the simultaneous execution of motor and cognitive tasks leads to decreased neurovascular coupling between fNIRS and EEG signals across theta, alpha, and beta frequency bands compared to single-task performance [18]. This degradation in coupling strength reflects the brain's limited capacity to maintain optimal neurovascular coordination when cognitive resources are divided. The observed reduction in NVC correlates with behavioral performance decrements, suggesting functional significance in coupling efficiency.
Pathological States
Various neurological conditions demonstrate altered NVC dynamics, providing insights into disease mechanisms. In retired rugby players with history of repetitive mild traumatic brain injury, fNIRS assessment reveals blunted hemodynamic responses during cognitive tasks compared to controls, indicating persistent neurovascular dysfunction [8]. Similarly, individuals with opiate addiction show desynchronized alpha rhythms and disrupted coupling between EEG oscillations and prefrontal hemodynamic responses [93]. These pathological alterations in NVC dynamics may underlie cognitive deficits and represent potential biomarkers for monitoring disease progression or treatment response.
Table 2: State-Dependent NVC Changes Across Conditions
| State/Condition | Neural Changes | Vascular Changes | Coupling Alterations |
|---|---|---|---|
| Sleep Inertia | Decreasing theta/beta ratio; Fluctuating alpha power [91] | Delayed thalamic BOLD response [91] | Time-varying lag in EEG-fMRI correlation [91] |
| Cognitive-Motor Dual-Task | Theta, alpha, beta power modifications [18] | Altered prefrontal oxygenation [18] | Reduced EEG-fNIRS correlation across frequency bands [18] |
| mTBI History | Frontal theta alterations | Reduced HbO increase in middle frontal gyrus [8] | Blunted hemodynamic response to cognitive tasks [8] |
| Opiate Addiction | Desynchronized lower alpha rhythms [93] | Decreased HbO-based connectivity in PFC [93] | Disrupted correlation between alpha oscillations and HbO [93] |
Experimental Protocols for Investigating Dynamic NVC
Simultaneous EEG-fMRI Protocol for Sleep Inertia
This protocol examines state-dependent NVC changes during the sleep-wake transition [91]:
Participant Preparation: Screen participants for sleep disorders using Pittsburgh Sleep Quality Index. Maintain consistent sleep-wake schedules for ≥3 days before experimentation, verified by wrist actigraphy. Restrict caffeine and alcohol on experiment day.
Data Acquisition: Nocturnal sleep occurs inside MRI scanner with MR-compatible EEG cap (32 electrodes). Acquire pre-sleep T1-weighted anatomical scan (6 min) followed by 5-min resting-state fMRI (eyes open). Allow up to 3 hours of sleep. Upon awakening, immediately conduct three consecutive 5-min resting-state fMRI sessions with 20-min interscan intervals.
Signal Processing: Preprocess fMRI data (realignment, segmentation, smoothing, normalization). Process EEG data with gradient artifact and ballistocardiogram removal. Compute EEG features including spectral slope (1-45 Hz), alpha-vigilance (alpha/[theta+delta]), and theta/beta ratio.
NVC Analysis: Calculate time-lagged correlations between EEG features and BOLD signals in regions of interest (thalamus, anterior cingulate, sensorimotor cortex). Identify optimal lags for peak correlation in each post-awakening session. Compare coupling strength and latency across sessions.
EEG-fNIRS Protocol for Cognitive-Motor Interference
This protocol assesses NVC dynamics during divided attention [18]:
Task Design: Implement three conditions: (1) Single motor task (grip force tracking), (2) Single cognitive task (number detection), (3) Cognitive-motor dual-task (simultaneous grip force tracking and number detection). Use block design with randomized condition order.
Multimodal Recording: Simultaneously record EEG (16+ channels according to 10-20 system) and fNIRS (covering prefrontal, motor cortices). Maintain synchronization between systems.
Data Processing: Apply task-related component analysis (TRCA) to both EEG and fNIRS signals to extract task-related components. For EEG, compute event-related spectral perturbations in theta, alpha, and beta bands. For fNIRS, convert optical density to oxygenated (HbO) and deoxygenated hemoglobin (HbR) concentrations.
NVC Quantification: Calculate correlation coefficients between EEG band powers and fNIRS HbO concentrations for each task condition. Statistically compare correlation strengths between single-task and dual-task conditions.
Multimodal EEG-fNIRS-TCD Protocol
This comprehensive protocol assesses NVC across arterial, capillary, and neuronal levels [62]:
Equipment Setup: Concurrently deploy EEG (16-channel 10-20 system), fNIRS (covering frontal, motor, parietal, occipital cortices), and TCD (insonating middle and posterior cerebral arteries). Synchronize all modalities with physiological monitoring (continuous blood pressure, capnography, heart rate).
Task Paradigm: Implement motor (finger tapping) and visual ("Where's Waldo?" search) tasks in block design. Include resting baseline periods between activation blocks.
Data Analysis: Perform time-frequency analysis on EEG data. Compute HbO and HbR concentration changes from fNIRS. Calculate cerebral blood velocity from TCD. Use cross-correlation analysis to quantify relationships between EEG features, fNIRS hemodynamics, and TCD flow velocities while controlling for systemic influences.
Diagram 2: State-Modulated Neurovascular Signaling Pathways
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Essential Research Resources for Dynamic NVC Studies
| Category | Specific Items | Research Function |
|---|---|---|
| Neuroimaging Equipment | MR-compatible EEG systems (32+ electrodes) [91] | Simultaneous EEG-fMRI acquisition without signal interference |
| fNIRS systems (multiple wavelengths) [8] [18] | Measures cortical hemoglobin concentration changes | |
| Transcranial Doppler (TCD) [62] | Assesses cerebral artery blood flow velocity | |
| Physiological Monitoring | Capnography [62] | Monitors end-tidal CO₂ for controlling respiratory influences |
| Continuous blood pressure monitoring [62] | Tracks systemic cardiovascular effects on cerebral circulation | |
| Actigraphy watches [91] | Verifies sleep-wake schedules before experimentation | |
| Experimental Paradigms | "Where's Waldo" visual search task [8] [62] | Engages sustained attention and visual processing |
| Cognitive-motor dual-tasks [18] | Creates divided attention conditions for NVC challenge | |
| Sleep inertia protocol [91] | Examines state transitions in neurovascular coupling | |
| Analysis Tools | Task-related component analysis (TRCA) [18] | Extracts task-relevant components from bimodal signals |
| Time-lagged correlation algorithms [91] | Identifies optimal temporal relationships in NVC | |
| Multimodal source power comodulation (mSPoC) [93] | Machine learning approach for EEG-fNIRS fusion |
Implications for fMRI and fNIRS Research
The dynamic nature of NVC necessitates methodological adjustments in functional neuroimaging research. First, vigilance states must be carefully monitored and controlled during imaging experiments, as fluctuations in alertness significantly impact neurovascular coupling [91]. Incorporating simultaneous EEG monitoring during fMRI studies provides valuable information about brain states that influence BOLD responses. Second, systemic physiological variables including blood pressure, end-tidal CO₂, and heart rate should be measured and incorporated as covariates in analyses, as these factors substantially confound neurovascular relationships [92] [62].
For clinical applications, dynamic NVC assessment offers promising biomarkers for various neurological conditions. The sensitivity of coupling measures to pathological states suggests potential utility in early detection, disease monitoring, and treatment evaluation [8] [93] [89]. Standardized protocols for probing NVC dynamics could enhance diagnostic precision and provide insights into disease mechanisms across neurodegenerative, cerebrovascular, and psychiatric disorders.
Future methodological developments should focus on computational models that incorporate state-dependent parameters for more accurate prediction of hemodynamic responses from neural activity [12]. Additionally, advancing multimodal integration frameworks will enable more comprehensive characterization of NVC across temporal and spatial scales, bridging microscopic cellular events with macroscopic neuroimaging signals.
Dynamic NVC represents a fundamental shift in how researchers conceptualize and study neurovascular relationships. Moving beyond static models to state-dependent frameworks acknowledges the brain's adaptive regulatory mechanisms and their profound influence on functional neuroimaging signals. The methodological approaches outlined in this technical guide provide researchers with tools to account for these dynamics in experimental design, data acquisition, and analysis. As the field advances, incorporating dynamic NVC perspectives will enhance the validity and interpretability of fMRI and fNIRS research, ultimately leading to more accurate models of brain function in health and disease.
Validation and Comparative Analysis: Integrating Multi-Modal Data for Robust NVC Assessment
In the investigation of human brain function, functional Near-Infrared Spectroscopy (fNIRS) and functional Magnetic Resonance Imaging (fMRI) have emerged as predominant hemodynamic-based neuroimaging techniques. Both methods rely on the principle of neurovascular coupling, the physiological link between neuronal activity and subsequent changes in cerebral blood flow and oxygenation [1]. However, each technique possesses distinct technical strengths and limitations, necessitating rigorous cross-validation against established neurophysiological and hemodynamic "gold standards" to ensure data accuracy and biological validity. Electroencephalography (EEG) provides direct measurement of neuronal electrical activity with millisecond temporal resolution, while Transcranial Doppler (TCD) offers non-invasive assessment of cerebral blood flow velocity in major arteries. This technical guide examines the methodological frameworks for validating fNIRS and fMRI findings against these established modalities, with particular emphasis on experimental protocols, quantitative comparisons, and integration strategies essential for researchers and drug development professionals conducting clinical neuroimaging research.
Physiological Foundations: Neurovascular Coupling and Hemodynamic Response
The Neurovascular Unit and Hemodynamic Response
Neurovascular coupling comprises complex cellular mechanisms that translate synaptic activity into regional changes in cerebral blood flow. During neural activation, both neurons and astrocytes release vasoactive factors including nitric oxide, prostaglandins, and epoxyeicosatrienoic acids that act on smooth muscle cells of arterioles to induce vasodilation [1]. This vascular response leads to a characteristic hemodynamic response function featuring a rapid increase in cerebral blood flow that typically peaks 2-6 seconds after stimulus onset, delivering oxygenated blood in excess of metabolic demands.
The resulting local hemodynamic changes form the basis for both fNIRS and fMRI measurements. fNIRS employs near-infrared light to measure relative concentration changes of oxygenated hemoglobin and deoxygenated hemoglobin in cortical tissue [45]. fMRI detects these changes indirectly through the Blood Oxygenation Level Dependent (BOLD) contrast, which reflects the magnetic susceptibility differences between oxygenated and deoxygenated blood [44]. This shared physiological origin enables direct comparison between modalities while highlighting their complementary measurement approaches.
Low-Frequency Oscillations as Cross-Modal Biomarkers
Recent research has identified low-frequency oscillations (LFOs) in the 0.01-0.2 Hz range as promising biomarkers for cross-modal validation studies. LFOs reflect intrinsic neurovascular coupling and demonstrate high correlation between fNIRS and EEG measurements [94]. Studies comparing healthy subjects with cerebral death patients have shown that LFO spectral power is significantly lower in cerebral death patients, confirming the cerebral origin of these signals and their value as indicators of viable neurovascular function [94].
Technical Comparison of Neuroimaging Modalities
Fundamental Measurement Principles and Capabilities
Table 1: Core Technical Specifications of Neuroimaging Modalities
| Parameter | fNIRS | fMRI | EEG | TCD |
|---|---|---|---|---|
| Primary Measurement | HbO₂/HbR concentration changes | BOLD signal (magnetic susceptibility) | Electrical potentials from pyramidal neurons | Cerebral blood flow velocity |
| Spatial Resolution | 1-3 cm (superficial cortex only) | 1-5 mm (whole brain) | 1-10 cm (poor localization) | Vessel-specific (large arteries) |
| Temporal Resolution | 0.1-10 Hz | 0.5-2 Hz (TR-dependent) | 0.001-0.5 s (ms precision) | 1-20 Hz |
| Penetration Depth | Superficial cortex (2-3 cm) | Whole brain | Superficial and deep sources | Large cerebral arteries |
| Portability | High (wearable systems available) | None (fixed scanner) | High (portable systems) | Moderate (bedside use) |
| Tolerance to Motion | Moderate (susceptible but correctable) | Poor (requires head immobilization) | Moderate (susceptible to EMG) | Good (allows some movement) |
| Population Flexibility | High (infants, children, implants) | Limited (claustrophobia, metal implants) | High (all populations) | High (all populations) |
| Key Artifact Sources | Systemic physiology, scalp blood flow | Cardiac, respiratory, motion | Ocular, muscle, line noise | Probe position, physiological variability |
fNIRS provides a favorable balance between spatial specificity and practical implementation for longitudinal studies and special populations. Its tolerance for movement and compatibility with metallic implants make it particularly suitable for drug development studies requiring repeated measurements [45] [44]. Conversely, fMRI remains the gold standard for whole-brain spatial localization but is limited by cost, accessibility, and the constrained scanner environment that may not reflect real-world brain function.
EEG offers direct measurement of neuronal population activity with millisecond temporal resolution, capturing neural dynamics that precede the hemodynamic response measured by fNIRS and fMRI [94]. TCD provides complementary information about macrovascular hemodynamics by measuring blood flow velocity in major cerebral arteries such as the middle cerebral artery, offering insights into global cerebral perfusion changes [95].
Quantitative Comparison of Hemodynamic Measurements
Table 2: Quantitative Parameters for Cross-Modal Correlation Studies
| Measurement Context | fNIRS-fMRI Correlation | fNIRS-EEG/TCD Correlation | Key Parameters |
|---|---|---|---|
| Prefrontal Activation During Cognitive Tasks | r = 0.75-0.95 for HbO₂-BOLD [45] | LFO-EEG coherence: 0.65-0.85 in healthy subjects [94] | HbO₂ concentration (μmol/L), BOLD signal (%), LFO power (dB) |
| Primary Motor Cortex Activation | Spatial correspondence: 84-92% for finger tapping [45] | TCD CBFv increase: 15-25% during motor activation | CBFv (cm/s), HbO₂ concentration (μmol/L), ERD/ERS in beta band (dB) |
| Visual Cortex Stimulation | HbO₂-BOLD delay: 1.5-2.5 s [44] | TCD PCA flow increase: 20-30% during visual stimulation | PCA flow velocity (cm/s), VEP latency (ms), HbO₂ peak (s) |
| Cerebral Autoregulation Assessment | COx-Mx correlation: r = 0.71-0.89 [96] | TCD autoregulation index: 4-8% change in CBFv per mmHg | COx index, Mx index, transfer function phase (degrees) |
| Patient Monitoring (Neurocritical Care) | Limited simultaneous studies; technical challenges | TCD vs. NIRS for vasospasm: 87% concordance [96] | Diastolic velocity (cm/s), Lindegaard ratio, HbO₂ trend |
Quantitative comparisons reveal generally strong correlations between fNIRS and fMRI hemodynamic measures, particularly for cortical regions accessible to both modalities. The relationship between fNIRS and EEG is more complex, requiring analysis of neurovascular coupling efficiency through metrics such as LFO-EEG coherence [94]. TCD provides essential validation for fNIRS measures of cerebral autoregulation, with the cerebral oximetry index demonstrating high correspondence with established TCD-based autoregulation measures [96].
Experimental Protocols for Cross-Validation Studies
Simultaneous fNIRS-fMRI Acquisition Protocol
Objective: To validate fNIRS hemodynamic measurements against the fMRI BOLD signal gold standard during controlled activation paradigms.
Equipment Requirements:
- MRI-compatible fNIRS system with fiber-optic cables
- 3T MRI scanner with compatible head coils
- fMRI presentation system for visual stimuli
- Response recording devices compatible with high magnetic fields
Procedure:
- Participant Preparation: Screen for MRI contraindications. Place fNIRS optodes according to the 10-20 system targeting regions of interest (e.g., prefrontal cortex for executive function tasks, motor cortex for finger tapping).
- Equipment Setup: Secure MRI-compatible fNIRS optodes using black cloth to prevent light leakage. Verify signal quality before positioning in scanner.
- Simultaneous Acquisition: Acquire fMRI BOLD signals (TR = 2000 ms, TE = 30 ms, voxel size = 3×3×3 mm³) while concurrently recording fNIRS data (sampling rate ≥ 10 Hz) during block design or event-related paradigms.
- Task Paradigm: Implement motor (finger tapping), cognitive (n-back task), or sensory (visual checkerboard) activation blocks (30s activation, 30s rest, 5-10 repetitions).
- Data Coregistration: Transform fNIRS optode locations to MRI coordinate space using fiducial markers for precise spatial correspondence.
Analysis Pipeline:
- Preprocess fNIRS data: convert raw intensity to optical density, motion artifact correction, bandpass filtering (0.01-0.2 Hz), conversion to HbO₂/HbR concentrations
- Preprocess fMRI data: motion correction, spatial smoothing, temporal filtering
- Extract time courses from both modalities for the same cortical regions
- Calculate correlation coefficients between HbO₂ and BOLD signal changes
- Perform general linear model analysis for both data types
This protocol capitalizes on the complementary strengths of both modalities: fMRI provides whole-brain coverage with high spatial resolution, while fNIRS offers higher temporal sampling of hemodynamic changes with lower susceptibility to movement artifacts [44].
fNIRS-EEG Protocol for Neurovascular Coupling Assessment
Objective: To investigate temporal relationships between electrical neural activity and hemodynamic responses using fNIRS and EEG.
Equipment Requirements:
- Simultaneous fNIRS-EEG system with integrated cap
- EEG amplifier with >64 channels
- fNIRS system with multiple wavelengths (690nm, 830nm)
- Stimulus presentation system
- Electrically shielded room (recommended)
Procedure:
- Montage Setup: Use integrated fNIRS-EEG cap with electrodes and optodes positioned according to the 10-20 system. Ensure optode-electrode spacing >1cm to minimize interference.
- Signal Quality Verification: Check EEG impedance (<10 kΩ) and fNIRS signal-to-noise ratio before beginning experimental tasks.
- Experimental Paradigm: Implement event-related designs with sensory (auditory tones, median nerve stimulation) or cognitive (oddball task) stimuli with random inter-stimulus intervals (8-12s).
- Data Acquisition: Record continuous EEG (sampling rate ≥ 500 Hz) and fNIRS (sampling rate ≥ 10 Hz) data throughout the experiment, marking stimulus events synchronously in both systems.
- Physiological Monitoring: Record heart rate and respiration to account for systemic physiological confounds in fNIRS signals.
Analysis Approach:
- Preprocess EEG data: filter (0.5-40 Hz), artifact removal, re-referencing
- Extract event-related potentials (ERP) for target stimuli
- Analyze fNIRS data to obtain HbO₂/HbR concentration changes time-locked to stimuli
- Calculate correlation between ERP components (e.g., N100, P300 amplitude) and HbO₂ response amplitude
- Perform time-frequency analysis of EEG to relate oscillatory power changes (event-related synchronization/desynchronization) to hemodynamic responses
This protocol enables direct investigation of neurovascular coupling efficiency by examining the temporal relationship between electrical brain responses (5-500 ms post-stimulus) and subsequent hemodynamic changes (2-8 s post-stimulus) [94].
fNIRS-TCD Protocol for Cerebral Autoregulation Monitoring
Objective: To validate fNIRS-derived measures of cerebral autoregulation against TCD as the clinical gold standard.
Equipment Requirements:
- TCD system with 2MHz pulsed-wave probes
- fNIRS system with bilateral forehead sensors
- Continuous blood pressure monitoring (Finapres or similar)
- Respiratory gas monitoring for CO₂ measurement (optional)
- Tilting table for orthostatic challenges
Procedure:
- Instrument Setup: Position TCD probes bilaterally on temporal windows to insonate middle cerebral arteries. Place fNIRS optodes on forehead to measure prefrontal cortex oxygenation.
- Baseline Recording: Collect 10 minutes of resting-state data with continuous TCD (CBFv), fNIRS (cerebral oximetry index), and blood pressure measurements.
- Autoregulation Challenges:
- Thigh Cuff Deflation: Inflate thigh cuffs to 30mmHg above systolic BP for 3 minutes, then rapidly deflate while recording cerebrovascular responses
- Orthostatic Stress: Implement head-up tilt to 60° for 5 minutes while monitoring CBFv and cerebral oxygenation
- CO₂ Reactivity: Measure responses to hypercapnia (5% CO₂ inhalation) and hyperventilation
- Data Synchronization: Ensure all physiological signals are recorded on a common timebase with synchronous event markers.
Analysis Methods:
- Calculate TCD-based autoregulation indices: mean flow velocity (MFV), pulsatility index (PI), transfer function analysis between BP and CBFv
- Compute fNIRS-based autoregulation measures: cerebral oximetry index (COx) as correlation between MAP and rSO₂, hemoglobin volume index (HVx)
- Determine CO₂ reactivity slopes for both TCD (%CBFv change/mmHg CO₂) and fNIRS (%rSO₂ change/mmHg CO₂)
- Compare dynamic autoregulation parameters between modalities using correlation analysis
This protocol is particularly valuable for drug development studies targeting cerebrovascular function, as it provides validated measures of cerebral autoregulation that can detect treatment effects on neurovascular function [96] [95].
The Researcher's Toolkit: Essential Solutions for Multimodal Studies
Table 3: Essential Research Reagents and Solutions for Cross-Validation Experiments
| Category | Specific Solution | Function/Application | Technical Notes |
|---|---|---|---|
| Integrated Hardware Platforms | Simultaneous fNIRS-fMRI systems | Enables direct temporal correlation of hemodynamic signals | Requires MRI-compatible fiber optics and shielding |
| Combined fNIRS-EEG caps | Standardized montages for spatial coregistration | Minimizes optode-electrode interference through physical design | |
| Software & Analytical Tools | AtlasViewer, NIRS-SPM, Homer2 | fNIRS data processing and anatomical coregistration | Enables mapping to standard brain space (MNI coordinates) |
| FieldTrip, SPM, FSL | Multimodal data integration and statistical analysis | Open-source platforms with specialized toolboxes | |
| Custom MATLAB/Python scripts | Calculation of cross-modal correlation metrics | Enables custom analysis pipelines for specific research questions | |
| Calibration & Validation Tools | Phantom testing systems | Validates fNIRS spatial sensitivity profiles | Uses tissue-simulating phantoms with known optical properties |
| 3D digitization systems | Precises optode/electrode localization | Critical for spatial accuracy in source reconstruction | |
| Physiological Monitoring | Continuous BP monitoring (Finapres) | Provides input for autoregulation calculations | Essential for TCD-fNIRS cerebral autoregulation studies |
| Capnography systems | Monitors end-tidal CO₂ for cerebrovascular reactivity | Accounts for CO₂-mediated vascular effects | |
| Motion tracking systems | Quantifies and corrects for movement artifacts | Especially important for naturalistic study designs |
Applications in Clinical Research and Drug Development
Multimodal validation approaches are particularly valuable in clinical trial contexts where establishing robust biomarkers of treatment response is essential. In neurodegenerative conditions such as Alzheimer's disease, combined fNIRS-fMRI approaches can track disease progression through changes in functional connectivity and neurovascular coupling efficiency [97]. For pharmaceutical studies targeting cerebrovascular function, TCD provides essential validation of fNIRS measures of cerebral autoregulation and CO₂ reactivity [96] [95].
In neurocritical care applications, fNIRS has emerged as a promising tool for continuous monitoring of patients with acute brain injury, with TCD serving as a periodic validation tool for assessing cerebral autoregulation status and identifying optimal blood pressure targets [96]. The correlation between fNIRS-derived cerebral oximetry index and TCD-based autoregulation measures enables continuous monitoring of autoregulation capacity, potentially guiding individualized therapy in neurocritical care settings [96].
Cross-validation of fNIRS and fMRI with established gold standards like EEG and TCD represents a methodological imperative for advancing cognitive neuroscience and neuropharmacology. The strong correlations demonstrated between modalities confirm the validity of fNIRS for measuring task-evoked cortical activation, particularly when combined with EEG's temporal precision and TCD's hemodynamic specificity. Future methodological developments will likely focus on standardized integration platforms, improved artifact correction algorithms, and multimodal biomarkers that combine the temporal precision of EEG with the spatial specificity of fMRI and the practical advantages of fNIRS. For drug development professionals, these validated multimodal approaches offer powerful tools for demonstrating treatment effects on neurovascular function across diverse clinical populations and settings.
Neurovascular coupling (NVC) is the fundamental physiological process whereby neuronal activity is linked to localized changes in cerebral blood flow, a mechanism critical for meeting the brain's dynamic metabolic demands [2]. This relationship forms the cornerstone for interpreting signals in non-invasive functional neuroimaging techniques such as functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS) [98] [2]. Whereas fMRI measures the blood oxygenation level-dependent (BOLD) signal and fNIRS measures concentration changes in oxygenated and deoxygenated hemoglobin, electroencephalography (EEG) provides a direct measure of the brain's electrical activity with millisecond temporal resolution [98] [99].
The simultaneous acquisition of EEG with either fNIRS or fMRI provides a powerful experimental paradigm to directly test and characterize NVC. This multimodal approach allows researchers to correlate the direct electrophysiological signals from EEG with the indirect hemodynamic responses captured by fMRI or fNIRS, thereby offering a more comprehensive window into brain function and the integrity of the neurovascular unit [98] [18] [34]. The ensuing technical guide details the experimental and analytical frameworks for employing these simultaneous modalities to advance our understanding of NVC in both healthy and diseased states.
Technical Foundations of Modalities
Core Principles and Complementarity
The rationale for multimodal integration rests on the complementary strengths and weaknesses of each technique, particularly regarding their spatial and temporal resolution, and the specific physiological phenomena they capture.
Table 1: Core Technical Characteristics of EEG, fNIRS, and fMRI
| Feature | EEG | fNIRS | fMRI |
|---|---|---|---|
| What It Measures | Electrical activity from postsynaptic potentials of cortical neurons [98] | Hemodynamic changes in oxygenated (HbO) and deoxygenated hemoglobin (HbR) concentrations [98] [8] | Blood Oxygenation Level-Dependent (BOLD) signal, a hemodynamic response [2] |
| Temporal Resolution | High (milliseconds) [99] | Low (seconds) [99] | Low (seconds) [98] |
| Spatial Resolution | Low (centimeter-level) [99] | Moderate for surface cortex [99] | High (millimeter-level) [98] |
| Key Strength | Direct neural activity, excellent temporal dynamics [98] | Good spatial localization for cortex, portable, silent operation [98] [32] | Whole-brain coverage, high spatial resolution [98] |
| Key Weakness | Poor spatial resolution, sensitive to artifacts [98] [99] | Limited to cortical surface, indirect and slow hemodynamic response [98] | Indirect and slow response, expensive, noisy environment [98] [32] |
The Neurovascular Coupling Hypothesis
The physiological basis for integrating EEG with hemodynamic modalities is the well-established neurovascular coupling mechanism [98] [2]. Upon neuronal activation, there is a release of neurotransmitters such as glutamate. This triggers signaling pathways within the neurovascular unit—comprising neurons, astrocytes, and vascular cells—leading to vasodilation of arterioles and an increase in local cerebral blood flow (CBF) [2]. This process, known as functional hyperemia, delivers oxygen and glucose to meet the energy demands of active neurons. The result is a local increase in oxyhemoglobin (HbO) and a decrease in deoxyhemoglobin (HbR), which is the source of the fNIRS and fMRI BOLD signals [98] [8] [2]. The hemodynamic response typically peaks 4-6 seconds after the neural event, creating a temporal lag that must be accounted for in analysis [32] [2].
Experimental Design and Protocols
Designing a robust experiment to test NVC requires careful consideration of the task paradigm, participant selection, and hardware integration.
Task Paradigms for NVC Investigation
Different cognitive and sensory tasks can be employed to elicit robust and measurable neural and hemodynamic responses.
Table 2: Exemplary Experimental Paradigms for NVC Studies
| Paradigm | Description | Measured Neural/Hemodynamic Correlates | Application in NVC |
|---|---|---|---|
| Auditory Intensity Dependence | Presentation of tones at varying intensity levels (e.g., 70-95 dB) [32]. | EEG: N1/P2 event-related potential (ERP) amplitudes [32].fNIRS/fMRI: HbO/HbR changes in auditory and prefrontal cortices [32]. | Tests the correlation between ERP amplitude and hemodynamic response magnitude as a function of stimulus intensity, a direct NVC metric [32]. |
| Cognitive-Motor Interference (CMI) | Simultaneous execution of a motor task (e.g., grip force) and a cognitive task (e.g., number detection) [18]. | EEG: Spectral power in theta, alpha, beta bands [18].fNIRS: Hemodynamic activity in prefrontal cortex [18]. | Investigates how divided attention affects the coupling strength between electrophysiological rhythms and hemodynamic responses [18]. |
| Sleep Inertia | Measuring brain state transitions from sleep to wakefulness with consecutive post-awakening sessions [34]. | EEG: Theta/Beta ratio, alpha-vigilance, spectral slope (1/f) [34].fMRI: BOLD signals in thalamus, anterior cingulate cortex (ACC) [34]. | Probes the state-dependence and dynamicity of NVC by examining time-lagged coupling between EEG and fMRI across different arousal levels [34]. |
| "Where's Wally" Visual Search | A visual search task to locate a specific character in a complex scene [8]. | fNIRS: HbO and HbR responses in the prefrontal cortex (DLPFC, OFC) [8]. | Serves as a standardized cognitive stressor to evoke a hemodynamic response for assessing NVC integrity, e.g., in concussed athletes [8]. |
Participant Considerations and Hardware Integration
Participant Groups: To investigate NVC pathology, studies often compare healthy controls (HCs) with clinical populations such as patients with Major Depressive Disorder (MDD) [9], a history of mild Traumatic Brain Injury (mTBI) [8], or neurodegenerative diseases [7]. Sample sizes in the cited studies typically range from 16 to 50 participants per group [18] [8] [9].
Simultaneous Recording Setup:
- EEG-fNIRS: The systems are compatible as they do not cause electromagnetic interference. Integration typically involves embedding fNIRS optodes into a standard EEG cap, using the international 10-20 system for co-registration [100]. Synchronization can be achieved via hardware triggers or a unified acquisition system [100].
- EEG-fMRI: This requires an MR-compatible EEG system equipped with specialized hardware to mitigate gradient and ballistocardiogram artifacts. Data synchronization is achieved using the scanner's trigger pulses [34].
Data Analysis Framework
Signal Preprocessing
Each data modality requires a dedicated preprocessing pipeline before integration.
- EEG Processing: Steps include filtering (e.g., 0.5-45 Hz), artifact removal (e.g., ocular, cardiac, muscle), and often re-referencing. For EEG-fMRI, robust gradient and ballistocardiogram artifact correction is essential [34].
- fNIRS Processing: Raw light intensity signals are converted to optical density and then to HbO and HbR concentration changes using the Modified Beer-Lambert Law [98]. Processing includes filtering to remove physiological noise (e.g., cardiac, respiratory) and motion artifacts [98] [18].
- fMRI Processing: Standard preprocessing includes realignment, slice-time correction, normalization to a standard space (e.g., MNI), and spatial smoothing [9] [34].
Analysis Methods for Neurovascular Coupling
Three primary methodological categories have been identified for concurrent fNIRS-EEG analysis, which can be extended to EEG-fMRI [98].
- EEG-informed fNIRS/fMRI Analysis: Features extracted from EEG (e.g., ERP amplitude, band power) are used as regressors in a general linear model (GLM) to predict the hemodynamic response [98] [32].
- fNIRS/fMRI-informed EEG Analysis: Activation areas identified from hemodynamic data are used as spatial constraints or regions of interest for source localization of EEG signals [98].
- Parallel fNIRS/fMRI and EEG Analysis: Data from both modalities are analyzed separately but then combined at the level of results, for instance, by calculating correlation coefficients between EEG spectral power and HbO/BOLD signal amplitudes across tasks or conditions [98] [18].
Advanced analytical frameworks are being developed to enhance the characterization of NVC. For instance, Task-Related Component Analysis (TRCA) can be applied to both EEG and fNIRS signals to extract components that are maximally reproducible across trials, improving the signal-to-noise ratio for subsequent correlation analysis [18]. Furthermore, to account for the dynamic nature of NVC, time-lagged correlation analysis can be employed to identify the specific delay at which the correlation between an EEG feature (e.g., alpha power) and a BOLD signal in a region of interest (e.g., thalamus) is strongest [34].
Key Findings and Clinical Applications
Simultaneous EEG-fNIRS and EEG-fMRI studies have yielded significant insights into NVC in both health and disease.
Table 3: Key Findings from Simultaneous NVC Studies
| Condition / Context | Key Findings | Implications for NVC |
|---|---|---|
| Cognitive-Motor Interference | A dual task led to decreased correlation (NVC strength) between fNIRS-HbO and EEG power in theta, alpha, and beta rhythms compared to single tasks [18]. | Cognitive interference and divided attention can weaken the temporal coupling between electrophysiological and hemodynamic activities [18]. |
| Auditory Processing | The amplitude of the N1/P2 ERP components increased with sound intensity. This was correlated with increased HbO in the auditory cortex, supporting a coupled response [32]. | Provides direct evidence of intensity-dependent NVC in the auditory system, validating the use of fNIRS in auditory paradigms [32]. |
| Sport-Related Concussion (mTBI) | Retired rugby players showed a blunted HbO response in the prefrontal cortex during a cognitive task compared to controls, indicating a reduced hemodynamic response for a given level of neural activity [8]. | Suggests a long-term impairment of NVC following repeated head injuries, potentially due to neurovascular unit dysfunction [8]. |
| Major Depressive Disorder (MDD) | First-episode, drug-naïve MDD patients showed reduced spatial correlation between neuronal activity (ALFF from fMRI) and cerebral blood flow (CBF from ASL) [9]. | Indicates neurovascular decoupling as a potential neuropathological mechanism in MDD, observable from the illness onset [9]. |
| Sleep Inertia | The time lag of the peak correlation between EEG alpha-vigilance and fMRI BOLD in the thalamus changed across post-awakening sessions, demonstrating dynamic NVC [34]. | Reveals that NVC is not static but varies with brain state and arousal level, challenging the assumption of a constant HRF [34]. |
The Scientist's Toolkit: Essential Research Reagents and Materials
This section details key materials, equipment, and software solutions essential for conducting simultaneous NVC studies.
Table 4: Essential Research Reagents and Materials for Simultaneous NVC Studies
| Item / Solution | Function / Description | Example Use Case |
|---|---|---|
| Integrated EEG-fNIRS Cap | A head cap with pre-defined placements for both EEG electrodes and fNIRS optodes, ensuring consistent co-registration [100]. | All simultaneous EEG-fNIRS studies for proper sensor placement and minimal cross-talk [18] [100]. |
| MR-Compatible EEG System | An EEG system with specialized amplifiers, electrodes, and cables designed to operate safely and effectively inside the MRI scanner, resistant to electromagnetic interference [34]. | Essential for all simultaneous EEG-fMRI studies to ensure patient safety and data quality [34]. |
| Synchronization Trigger Box | Hardware device that sends TTL pulses to synchronize the clocks of different acquisition systems (e.g., EEG and fNIRS/fMRI) with millisecond precision [100]. | Critical for temporal alignment of data streams in all simultaneous recording setups [100] [34]. |
| Abrasive Electrode Paste (e.g., ABRALYT HiCl) | Used to prepare the scalp and lower impedance at EEG electrode sites (< 15 kΩ), ensuring high-quality electrical signal acquisition [34]. | Standard preparation for EEG in both EEG-fNIRS and EEG-fMRI experiments [34]. |
| Task-Related Component Analysis (TRCA) Algorithm | A data-driven algorithm that extracts task-related components by maximizing inter-trial covariance, enhancing signal quality for NVC analysis [18]. | Used to improve the reproducibility and discriminability of EEG and fNIRS features before correlation analysis [18]. |
| Arterial Spin Labeling (ASL) MRI Sequence | An MRI technique that uses magnetically labeled arterial blood water as an endogenous tracer to quantitatively measure cerebral blood flow (CBF) [9]. | Used in conjunction with BOLD-fMRI to provide a direct measure of CBF for spatial/temporal NVC analysis [9]. |
Simultaneous EEG-fNIRS and EEG-fMRI provide a direct and powerful means to test the mechanisms and integrity of neurovascular coupling. These multimodal approaches overcome the limitations of single-modality imaging by combining the superior temporal resolution of EEG with the localized hemodynamic information from fNIRS and fMRI. The experimental and analytical frameworks outlined in this guide have begun to reveal that NVC is a dynamic process, sensitive to cognitive state, and impaired in a range of neurological and psychiatric disorders. Future advancements will likely come from improved hardware integration, standardized analytical pipelines, and the application of machine learning to model the complex relationship between neural electrical activity and its vascular consequences.
- Basic principles: Introduces fMRI and fNIRS fundamentals and neurovascular coupling.
- Technical specifications: Compares resolution metrics in a table.
- Methodological approaches: Details experimental protocols and integration methods.
- Advanced applications: Explores clinical and research uses with a reagents table.
- Visualization: Includes DOT scripts for signaling pathways and workflows.
Comparative Analysis of Spatial and Temporal Resolution: fMRI vs. fNIRS
The complex functional organization of the human brain requires multimodal neuroimaging approaches to fully elucidate its intricate operations. Functional magnetic resonance imaging (fMRI) and functional near-infrared spectroscopy (fNIRS) have emerged as two complementary hemodynamic-based neuroimaging techniques that leverage the principle of neurovascular coupling—the fundamental process where neural activity triggers localized changes in cerebral blood flow, volume, and oxygenation [36] [32]. When neurons become active, they initiate a complex cascade of vascular events that ultimately leads to an increased oxygen supply disproportionate to the actual oxygen consumption, resulting in characteristic changes in hemoglobin oxygenation states that both techniques detect through different physical mechanisms [32] [8].
fMRI measures brain activity indirectly through the blood-oxygen-level-dependent (BOLD) signal, which exploits the different magnetic properties of oxygenated and deoxygenated hemoglobin. When brain regions become active, the local vascular response typically leads to a decrease in deoxygenated hemoglobin concentration, which alters the magnetic susceptibility of blood and consequently affects the MRI signal [45]. In contrast, fNIRS directly measures hemoglobin concentration changes by transmitting near-infrared light (650-950 nm) through the scalp and skull and detecting the attenuated light that emerges from the tissue. The different absorption spectra of oxygenated hemoglobin (HbO) and deoxygenated hemoglobin (HbR) at various wavelengths enable the calculation of relative concentration changes for both chromophores [36] [45]. While both techniques ultimately reflect the same underlying neurovascular processes, their distinct measurement principles, operational requirements, and physical constraints result in significantly different spatial and temporal resolution characteristics that determine their appropriate applications in neuroscience research and clinical practice.
Technical Specifications and Resolution Metrics
Spatial Resolution Characteristics
The spatial resolution of neuroimaging techniques determines their ability to localize neural activity and distinguish between adjacent functional regions. fMRI provides comprehensive brain coverage with exceptional spatial precision, typically achieving millimeter-level resolution (1-3 mm) that enables visualization of both cortical and deep subcortical structures, including the hippocampus, amygdala, and thalamus [36] [45]. This whole-brain coverage facilitates the simultaneous examination of multiple brain networks and their interactions, making fMRI particularly advantageous for investigating complex neural systems underlying psychiatric and neurological disorders [36].
In contrast, fNIRS exhibits significantly constrained spatial resolution (typically 1-3 centimeters) due to the rapid scattering of near-infrared light as it passes through biological tissues [36] [45]. This limited resolution restricts precise localization of brain activity and presents challenges for differentiating adjacent functional regions. Furthermore, fNIRS is confined to monitoring superficial cortical regions due to the limited penetration depth of near-infrared light (approximately 1-1.5 cm), rendering it unsuitable for investigating subcortical structures [36] [45]. However, recent technological advances have demonstrated that high-density fNIRS arrays with overlapping, multidistance channels can improve spatial resolution and localization accuracy. These high-density configurations enhance sensitivity and reduce partial volume blurring, with some studies showing that their sensitivity approaches that of fMRI [101].
Temporal Resolution Profiles
Temporal resolution refers to the ability of a neuroimaging technique to track dynamic changes in brain activity over time. fNIRS demonstrates superior temporal resolution compared to fMRI, often achieving millisecond-level precision with sampling rates typically around 10 Hz [36] [102]. This enhanced temporal capability allows fNIRS to capture more rapid neural dynamics and better distinguish physiological artifacts (e.g., cardiac cycles ~1 Hz, respiratory activities ~0.3 Hz) from the signals of interest [102].
Conversely, fMRI experiences notable temporal constraints due to the inherent characteristics of the hemodynamic response. The BOLD signal typically lags behind neural activity by 4-6 seconds, with a sampling rate generally ranging from 0.33 to 2 Hz [36] [103]. This limited temporal resolution stems from the blurred intrinsic hemodynamic response and finite signal-to-noise ratio, restricting fMRI's ability to resolve rapidly evolving neural processes [103]. Nevertheless, studies have demonstrated that fMRI can distinguish time courses in motor areas with differences as small as 2 seconds when examining spatially distinct regions, suggesting that temporal resolution varies across brain areas and experimental paradigms [103].
Table 1: Comprehensive Comparison of Technical Specifications Between fMRI and fNIRS
| Parameter | fMRI | fNIRS |
|---|---|---|
| Spatial Resolution | 1-3 mm (high) | 1-3 cm (moderate) |
| Temporal Resolution | 0.33-2 Hz (slow) | Up to 10 Hz (typically 0.1-0.2 Hz) |
| Penetration Depth | Whole brain (deep structures accessible) | Superficial cortex (1-1.5 cm) |
| Measured Parameters | BOLD signal (reflects Δ[HbR]) | Δ[HbO] and Δ[HbR] concentrations |
| Primary Physiological Basis | Neurovascular coupling | Neurovascular coupling |
| Typical Sampling Rate | 0.5-2 Hz | ~10 Hz |
Methodological Approaches and Experimental Protocols
fMRI Experimental Paradigms and Processing
fMRI experimental design requires careful consideration of the hemodynamic response latency when constructing task paradigms. Block designs, which alternate between task and rest conditions in extended periods (typically 20-30 seconds), leverage the sustained hemodynamic response to maximize detection power. Event-related designs, which present discrete trials with shorter durations, allow for analysis of individual hemodynamic responses to specific stimuli but typically yield lower signal-to-noise ratios [36]. For auditory paradigms specifically, fMRI faces significant challenges due to the substantial background noise generated by gradient coils during scanning, which can reach levels that interfere with auditory stimulation and perception [32].
Processing fMRI data involves multiple steps including motion correction, spatial smoothing, temporal filtering, and statistical analysis. The BOLD signal is often modeled using a canonical hemodynamic response function (HRF) to account for the characteristic delay and dispersion of the neurovascular response. Recent advances have focused on characterizing regional variations in hemodynamic timing, which can serve as important biomarkers of cerebrovascular health and potential confounds in functional connectivity analyses [104]. Studies have demonstrated that hemodynamic timing is more robustly characterized when larger systemic vascular responses are evoked by breathing challenges compared to spontaneous fluctuations present in resting-state data [104].
fNIRS Experimental Protocols and Signal Quality
fNIRS experimental implementation must address several unique methodological considerations. Proper optode placement is critical for targeting specific cortical regions, with techniques such as 3D digitization and brain mapping software helping to ensure accurate positioning based on anatomical landmarks [45] [102]. The development of high-density arrays with overlapping source-detector pairs has significantly improved spatial specificity and signal quality, though at the cost of increased setup complexity and computational requirements [101].
Signal quality assurance in fNIRS requires rigorous preprocessing to address multiple contamination sources. The scalp-coupling index serves as a valuable metric for assessing signal quality by evaluating the presence of cardiac pulsations in the fNIRS data [63]. Additional quality control measures include the identification and exclusion of bad channels, motion artifact correction, and separation of cerebral signals from superficial physiological noise using short-separation channels [63] [102]. Recent research has revealed that signal quality can vary significantly based on participant characteristics and task demands, with studies showing that task type influences data quality, and that fNIRS signals were generally worse for Black women compared to Black men and White individuals regardless of gender, highlighting the need for hardware improvements to ensure equity in fNIRS research [63].
Multimodal Integration Approaches
The complementary strengths of fMRI and fNIRS have motivated the development of integrated approaches that combine both modalities. There are two primary integration methods: synchronous acquisition, where both signals are recorded simultaneously, and asynchronous acquisition, where data are collected separately but under comparable conditions [36]. Synchronous acquisition enables direct temporal correlation between fMRI and fNIRS signals but presents technical challenges such as electromagnetic interference in the MRI environment [36].
Advanced integration frameworks have been developed to facilitate combined analysis, including surface-based approaches that project both fNIRS and fMRI data onto cortical surface meshes derived from anatomical MRI. This method enables direct comparison within the same anatomical space and has demonstrated moderate to substantial spatial agreement (Dice coefficient range: 0.43-0.69) and moderate to strong temporal correlations (Pearson's r: 0.79-0.85 for BOLD vs. HbO) between modalities [105]. These integration techniques are particularly valuable for applications requiring ecological validity, such as longitudinal monitoring of brain activity before and after rehabilitation [105].
Advanced Applications and Research Implications
Clinical and Cognitive Applications
The complementary resolution profiles of fMRI and fNIRS have enabled their application across diverse clinical and research domains. fMRI remains the gold standard for precise spatial localization in basic cognitive neuroscience, clinical diagnostics, and pre-surgical planning, where millimeter-level precision is essential [36] [45]. Its whole-brain coverage facilitates comprehensive mapping of functional networks and their alterations in neurological and psychiatric conditions such as Alzheimer's disease, depression, and autism spectrum disorder [36].
fNIRS has found particular utility in scenarios where fMRI's practical limitations are prohibitive. Its mobility and resistance to motion artifacts make it ideally suited for studying naturalistic behaviors, including social interactions, rehabilitation exercises, and real-world cognitive tasks [36] [45]. The portability of fNIRS systems enables brain imaging in various settings beyond traditional laboratories, including bedside monitoring, field studies, and investigations involving special populations such as infants, children, and individuals with motor impairments who may have difficulty remaining still in the MRI scanner [36] [45]. Additionally, fNIRS has emerged as a valuable tool for assessing neurovascular coupling impairments in conditions such as sport-related concussion, with studies demonstrating reduced cerebral hemodynamic responses in retired athletes with a history of mild traumatic brain injury compared to controls [8].
Real-Time Applications and Future Directions
The temporal advantages of fNIRS have positioned it as a promising modality for real-time neuroimaging applications such as neurofeedback and brain-computer interfaces (BCIs). fNIRS provides an optimal balance between spatial specificity and mobility for these applications, with typical sampling rates (~10 Hz) sufficient for tracking the hemodynamic response while offering greater portability than fMRI [102]. Real-time fNIRS applications face unique methodological challenges, including the need for robust online processing pipelines that can maintain consistent calculation times while ensuring that incoming data reflect true underlying brain activity rather than noise or artifacts [102].
Future developments in both technologies will likely focus on addressing current limitations while enhancing integration approaches. For fNIRS, hardware innovations including MRI-compatible probes, improved depth resolution through advanced reconstruction algorithms, and machine learning-driven data analysis represent active areas of development [36]. For fMRI, efforts continue to enhance temporal resolution through accelerated acquisition sequences and to better characterize and account for regional variations in hemodynamic timing [104]. The complementary nature of these techniques ensures that their continued integration will advance our understanding of brain function across laboratory and real-world settings.
Table 2: Essential Research Reagents and Tools for fMRI-fNIRS Research
| Tool/Reagent | Primary Function | Application Context |
|---|---|---|
| High-Density fNIRS Arrays | Improved spatial localization through overlapping measurements | Cognitive neuroscience, clinical studies |
| Short-Separation Channels | Regression of superficial physiological noise | Signal quality improvement in fNIRS |
| 3D Digitization Systems | Precise optode localization relative to anatomy | Spatial registration in fNIRS |
| AtlasViewer Software | Brain mapping based on standard anatomical templates | fNIRS data analysis and visualization |
| QT-NIRS Toolbox | Quality testing and signal quality metrics | fNIRS data quality assurance |
| Surface-Based Integration Algorithms | Multimodal data fusion in common anatomical space | Combined fMRI-fNIRS analysis |
Visualization of Neurovascular Coupling and Technical Integration
Neurovascular Coupling Pathway
The following diagram illustrates the fundamental neurovascular coupling process that underlies both fMRI and fNIRS signal generation:
Diagram 1: Neurovascular Coupling Pathway. This diagram illustrates the sequential biological process linking neural activity to hemodynamic changes measured by both fMRI and fNIRS.
Multimodal Integration Workflow
The following workflow depicts the process for integrating fMRI and fNIRS data collection and analysis:
Diagram 2: Multimodal fMRI-fNIRS Integration Workflow. This diagram illustrates the sequential process for combining data from both neuroimaging modalities to leverage their complementary strengths.
This comparative analysis demonstrates that fMRI and fNIRS offer complementary rather than competing capabilities for studying brain function through neurovascular coupling. fMRI provides unparalleled spatial resolution and whole-brain coverage, making it ideal for precise localization of neural activity and investigation of deep brain structures. Conversely, fNIRS offers superior temporal resolution, portability, and tolerance to motion artifacts, enabling brain imaging in naturalistic settings and with populations inaccessible to fMRI. The ongoing development of multimodal integration approaches, including surface-based analysis frameworks and simultaneous acquisition protocols, continues to enhance our ability to leverage the strengths of both techniques. As both technologies evolve, their synergistic application promises to advance our understanding of brain function across laboratory and real-world contexts, ultimately bridging critical gaps in spatial and temporal characterization of neural activity.
Neurovascular coupling (NVC) is the fundamental biological mechanism that dynamically links local neural activity to subsequent changes in cerebral blood flow (CBF), ensuring the brain's energy demands are met during cognitive and motor tasks [7] [12]. This process is orchestrated by the neurovascular unit (NVU), a functional complex comprising neurons, astrocytes, vascular cells, and pericytes [7]. In recent years, NVC dysfunction has been increasingly implicated in the pathophysiology of various neurodegenerative diseases, including Alzheimer's disease (AD) and related dementias [7] [106]. In conditions like AD, pathological hallmarks such as amyloid-β deposition adversely affect endothelial function and pericyte signaling, thereby compromising the NVU's ability to match blood flow to neural demand [7]. This impairment in NVC leads to disrupted cerebral blood flow regulation, contributing to metabolic stress, accumulation of toxic proteins, and ultimately, neuronal injury [7]. Consequently, the validation of NVC impairments has emerged as a critical area of investigation, providing not only potential biomarkers for early diagnosis but also promising therapeutic targets for mitigating disease progression [7] [107]. This technical guide synthesizes current methodologies and findings from key case studies that validate NVC impairments across neurodegenerative disease models, with particular emphasis on integrated fMRI and fNIRS approaches.
Case Study 1: fNIRS-Based NVC Impairment in Mild Cognitive Impairment
A 2024 study investigated NVC and functional connectivity (FC) alterations in patients with mild cognitive impairment (MCI), a recognized prodromal stage of dementia [107]. The research aimed to determine characteristic functional determinants of MCI by testing the hypothesis that impaired NVC responses and decreased FC would effectively classify MCI with high feature importance. Furthermore, the study explored the relationship between plasma levels of cerebrovascular endothelial extracellular vesicles (CEEVs) and cerebrovascular small vessel ischemic burden in MCI participants [107]. This comprehensive approach sought to bridge existing gaps in mechanistic understanding of MCI development and progression through the lens of cerebrovascular pathology.
Detailed Methodological Protocol
- Participant Cohort: Community-dwelling older adults and participants with MCI were recruited for this cross-sectional observational study. Inclusion criteria consisted of age >50 years, Clinical Dementia Rating (CDR) equal to 0.5 and/or Montreal Cognitive Assessment (MoCA) <26 and ≥19 for the MCI group. Control participants (CN) required MoCA ≥26 and no clinical diagnosis of cognitive impairment [107].
- fNIRS Acquisition: Participants underwent functional near-infrared spectroscopy (fNIRS) recording during administration of the n-back working memory paradigm. The optical system measured concentration changes in oxygenated hemoglobin (HbO) and deoxygenated hemoglobin (HbR) in the prefrontal cortex [107].
- Hemodynamic Response Quantification: The fNIRS signal was processed to extract hemodynamic response functions during task performance. NVC was quantified as the magnitude of the HbO response to cognitive activation, while FC was assessed through statistical relationships between low-frequency oscillations in the HbO signal across different brain regions [107].
- Extracellular Vesicle Analysis: Blood samples were collected from participants, and plasma levels of extracellular vesicles (EVs) were assessed using small-particle flow cytometry. Specific attention was given to cerebrovascular endothelial EVs (CEEVs) as potential markers of active cerebrovascular disease [107].
- Structural MRI and White Matter Hyperintensity Assessment: T2-weighted FLAIR 1.5 Tesla MRI scans were utilized for ordinal quantification of subcortical deep white matter lesions using the Fazekas scale, graded by an experienced vascular neurologist blinded to functional neuroimaging and EV data [107].
- Statistical Analysis and Machine Learning Classification: Random Forest classification was employed to determine the feature importance of NVC, FC, and CEEVs in distinguishing MCI from control participants, alongside demographic and comorbidity factors [107].
Key Findings and Quantitative Results
The study yielded several significant findings validating NVC impairment in MCI:
- NVC and FC Reduction: NVC and FC were significantly decreased in MCI compared to cognitively normal controls, with the most prominent deficits observed in the left dorsolateral prefrontal cortex (LDLPFC) during working memory tasks [107].
- CEEV Elevation: The ratio of CEEVs to total endothelial EVs was significantly increased in patients with MCI compared to CN. This ratio demonstrated a strong correlation with structural MRI evidence of small vessel ischemic damage in the MCI group [107].
- Predictive Power: In random Forest classification, LDLPFC NVC, CEEV ratio, and LDLPFC FC demonstrated the highest feature importance for predicting MCI diagnosis, outperforming traditional demographic and comorbidity factors [107].
Table 1: Key Quantitative Findings from MCI NVC Study
| Parameter | Finding | Significance |
|---|---|---|
| LDLPFC NVC | Significantly decreased in MCI | Primary feature in MCI classification |
| LDLPFC FC | Significantly decreased in MCI | Loss of compensatory mechanism |
| CEEV Ratio | Significantly increased in MCI | Correlated with small vessel ischemic damage |
| Classification Accuracy | High with NVC, CEEVs, and FC | Superior to demographic/comorbidity models |
Diagram 1: Experimental workflow for MCI case study
Case Study 2: NIRS of Cerebellar NVC in Motor Activation Tasks
Study Objectives and Technical Challenges
A 2025 study addressed the technical challenges of measuring cerebellar NVC during motor tasks, an area previously limited by motion artifacts in fMRI and the cerebellar depth relative to the cortical surface [108]. The research aimed to establish the efficacy of NIRS in evaluating cerebellar function during standardized clinical motor tasks in healthy subjects by employing two novel protocols. A secondary objective determined whether the methods could effectively differentiate cerebellar activity from corticospinal motor activity for each task [108]. This validation is particularly important for neurodegenerative diseases like Parkinson's and multiple system atrophy where cerebellar dysfunction contributes to motor symptoms.
Experimental Protocols and Technical Specifications
- Participant Setup: All subjects were seated comfortably with eyes closed in a dark, sound-attenuated room to eliminate confounding visual and auditory stimuli. Both hands were gently supported in the extended position, with subjects receiving verbal cues to start and stop performing each task [108].
- Motor Task Paradigms:
- Task 1 (Cerebellar): Subjects performed alternating pronation and supination movements with one hand clapping onto the inactive opposite hand. This bedside maneuver assesses diadochokinesia, a fundamental cerebellar function [108].
- Task 2 (Corticospinal): Subjects performed finger-thumb opposition from index to little finger repeatedly at maximal velocity. This action is primarily mediated by the corticospinal system up to the sensorimotor cortex [108].
- Experimental Sequence: After an initial 2-minute rest for baseline stabilization, participants performed tasks at maximal velocity with simultaneous fNIRS recording for 3 minutes, followed by a 3-minute rest period. Subjects were cued to perform the motor task on the opposite hand, with the cycle repeated three times total for each side [108].
- NIRS Configurations:
- Protocol 1: Utilized 5 optodes (4 sources, 1 detector) spaced 3 cm apart with single-tipped LED light sources and single-tipped fiber-optic detectors, yielding 4 recording channels [108].
- Protocol 2: Employed 10 optodes (2 sources, 8 detectors) spaced 2 cm apart with single-tipped LED light sources and dual-tipped fiber-optic detectors, yielding 8 recording channels [108].
- Data Analysis: Researchers measured mean baseline-to-peak changes for HbO and HbR in real time during each task, along with time-to-peak concentration values. Stable recordings were defined as waveforms showing distinct onset, maximum/minimum peaks within the recording period across all channels [108].
Key Outcomes and Technical Validation
- Protocol Superiority: Protocol 2 achieved stable recordings in 100% of subjects compared to only 50% with Protocol 1, demonstrating the enhanced feasibility of the optimized configuration for cerebellar assessment [108].
- Cerebellar Specificity: Protocol 2 detected significantly increased neurovascular coupling activity specifically during Task 1 (cerebellar task) compared to Task 2 (corticospinal task), confirming the ability to isolate cerebellar NVC from inherent corticospinal activity [108].
- Quantitative Hemodynamic Changes: The optimized protocol provided robust quantification of HbO and HbR amplitude changes and temporal dynamics during cerebellar-specific motor activation, establishing a validated approach for detecting potential NVC impairments in cerebellar neurodegenerative pathologies [108].
Table 2: NIRS Protocol Performance Comparison for Cerebellar NVC Assessment
| Parameter | Protocol 1 | Protocol 2 |
|---|---|---|
| Optode Configuration | 4 sources, 1 detector | 2 sources, 8 detectors |
| Channel Count | 4 recording channels | 8 recording channels |
| Optode Spacing | 3 cm apart | 2 cm apart |
| Detector Type | Single-tipped | Dual-tipped |
| Recording Stability | 50% of subjects | 100% of subjects |
| Cerebellar Specificity | Not achieved | Successfully achieved |
Common Mechanisms of NVC Dysfunction in Neurodegeneration
Across neurodegenerative diseases, common mechanistic themes emerge in NVC dysfunction, highlighting shared pathophysiological pathways despite etiological differences. In Alzheimer's disease, amyloid-β deposition directly impairs endothelial function and pericyte signaling within the neurovascular unit, compromising the brain's ability to regulate blood flow in response to neural activity [7]. Cerebrovascular endothelial dysfunction is further evidenced by increased levels of cerebrovascular endothelial extracellular vesicles (CEEVs) in MCI patients, which correlate strongly with small vessel ischemic damage observed on structural MRI [107]. Neuroinflammatory processes drive significant alterations in cellular biomechanics and extracellular matrix deposition, particularly in regions like the optic nerve head in glaucoma, creating a vicious cycle of impaired perfusion and neuronal injury [106]. Additionally, pathological protein aggregations characteristic of various neurodegenerative diseases (Aβ and tau in AD, α-synuclein in Parkinson's) directly disrupt neurovascular signaling pathways, while metabolic impairments and oxidative stress further compromise vascular function, ultimately leading to the uncoupling of neural activity from cerebral blood flow regulation [7] [106].
Diagram 2: NVC signaling pathways and disruption sites in neurodegeneration
Advanced Multimodal Integration: fMRI-fNIRS Synergy
The integration of multiple neuroimaging modalities has emerged as a powerful approach to overcome the limitations of individual techniques, with combined fMRI-fNIRS applications particularly advancing NVC investigation in neurodegeneration. This synergistic combination capitalizes on fMRI's high spatial resolution (localizing brain activity with millimeter-level precision across cortical and subcortical structures) alongside fNIRS's superior temporal resolution (capturing hemodynamic dynamics with millisecond-level precision) and operational flexibility in naturalistic settings [36]. The integration methodologies have evolved along two primary pathways: synchronous data acquisition, where both modalities record simultaneously to enable direct correlation of spatial and temporal hemodynamic patterns; and asynchronous acquisition, where data are collected separately but co-registered for complementary analysis [36]. This multimodal approach has proven particularly valuable in clinical neurodegeneration research, where fNIRS's portability enables bedside monitoring of patients alongside the detailed structural and functional insights provided by fMRI [36]. Furthermore, the combined use of similar hemodynamic-based techniques provides crucial validation of fNIRS measurements against the established gold standard of fMRI, strengthening the reliability of NVC impairment findings in patient populations [36].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Research Reagents and Materials for NVC Impairment Studies
| Item | Function/Application | Technical Specifications |
|---|---|---|
| fNIRS Systems | Measures HbO/HbR concentration changes during neural activation | NIRSPORT systems; 760 nm & 850 nm wavelengths; modified Beer-Lambert law [108] |
| fMRI Platforms | High-spatial resolution BOLD signal mapping | 1.5T/3T scanners; BOLD contrast; whole-brain coverage including subcortical structures [36] |
| Extracellular Vesicle Analysis | Quantification of cerebrovascular endothelial EVs as disease biomarkers | Small-particle flow cytometry; CEEV-specific surface markers [107] |
| Cognitive Task Paradigms | Standardized neural activation protocols | n-back working memory tasks; clinical motor tasks (diadochokinesia assessment) [107] [108] |
| Structural MRI Sequences | Assessment of structural correlates (WMH, atrophy) | T2-weighted FLAIR sequences; Fazekas scale for small vessel disease [107] |
| Data Fusion Algorithms | Integration of multimodal neuroimaging data | Machine learning approaches (Random Forest); synchronous/asynchronous integration methods [107] [36] |
The validation of NVC impairments through carefully designed case studies has established neurovascular dysfunction as a fundamental component of neurodegenerative disease pathophysiology rather than merely a secondary consequence. The integration of multimodal approaches, particularly fNIRS and fMRI, provides a powerful framework for quantifying these impairments with both spatial precision and temporal resolution. Future research directions will likely focus on leveraging artificial intelligence and machine learning to enhance the predictive power of NVC biomarkers, integrating multi-omics analyses to elucidate molecular pathways, and developing high-resolution imaging techniques to further clarify NVC mechanisms in health and disease [7]. These advances will promote interdisciplinary translation and facilitate breakthroughs in both neuroscience and therapeutic development for neurodegenerative disorders, potentially enabling interventions that preserve neurovascular function early in the disease course.
The interpretation of functional neuroimaging data, a cornerstone of modern neuroscience and drug development, rests entirely on a critical physiological process: neurovascular coupling (NVC). NVC describes the intricate mechanism by which neural activity triggers dynamic changes in local cerebral blood flow and oxygenation [39] [4]. For decades, researchers have relied on functional Magnetic Resonance Imaging (fMRI) and functional Near-Infrared Spectroscopy (fNIRS) to non-invasively probe brain function, with the Blood Oxygenation Level-Dependent (BOLD) signal serving as a proxy for this underlying neural activity [109] [3]. However, the relationship between neural firing and the subsequent hemodynamic response is profoundly complex and nonlinear, governed by a cascade of cellular signaling events. Traditional analytical approaches often treat this relationship as a statistical black box, limiting the biological interpretability of findings [4] [3].
The emergence of multimodal artificial intelligence (AI) presents a paradigm shift. By simultaneously processing and learning from diverse data types—such as electrophysiological recordings (MEG/EEG), hemodynamic signals (fMRI/fNIRS), and clinical metadata—multimodal AI models can disentangle the fundamental mechanisms of NVC [109] [110]. This integration is transforming neuroimaging from a purely correlative tool into a platform for mechanistic inference. The global multimodal AI market is projected to reach a valuation of $10.89 billion by 2030, driven by its transformative potential in technology and healthcare [111]. This technical guide explores how advanced computational frameworks, including dynamic causal modelling and deep learning, are being leveraged to create a unified, quantitative understanding of neurovascular function, thereby enhancing the precision of biomarker discovery and accelerating therapeutic development for neurological disorders.
The Current Landscape of Neurovascular Coupling Research
Understanding NVC is essential because it forms the biological basis for signals measured by fMRI and fNIRS. A comprehensive understanding of its mechanisms is critical for accurately interpreting this data, especially in the context of disease states where the coupling may be impaired.
Key Physiological Mechanisms and Signaling Pathways
At its core, NVC is mediated by synaptic activity that triggers the release of vasoactive molecules from various neural cells. These molecules act on vascular smooth muscle cells and pericytes to modulate cerebral blood flow (CBF) and volume (CBV) [4] [3]. The central signaling pathways involve:
- Pyramidal Neurons: Activation leads to calcium influx, promoting the synthesis of arachidonic acid (AA) via phospholipase A2 (PLA2), which is subsequently metabolized to the vasodilator prostaglandin E2 (PGE2) via cyclooxygenase-2 (COX-2) [4].
- GABAergic Interneurons: Specific sub-populations contribute distinct temporal components to the vascular response. NO-interneurons mediate the first rapid dilation, while NPY-interneurons are responsible for the post-peak undershoot in the hemodynamic response [3].
- Astrocytes: These glial cells synthesize several vasoactive messengers, including PGE2 (via COX-1), epoxyeicosatrienoic acid (EET), and 20-HETE, which collectively fine-tune the vascular response [4].
The following diagram illustrates the core cellular signaling pathways involved in neurovascular coupling, showing how neuronal activity triggers a cascade of vasoactive signals.
Quantitative Research Trends and Focal Points
Scientometric analysis of the past three decades (1996-2025) of NVC research reveals a rapidly evolving field. An analysis of 2,047 articles from the Web of Science Core Collection highlights dominant research trends and the growing importance of non-invasive imaging techniques [39].
Table 1: Key Research Metrics and Trends in Neurovascular Coupling (1996-2025) [39]
| Metric | Findings | Implications |
|---|---|---|
| Total Publications | 2,047 articles | Sustained and growing academic interest in the field. |
| Leading Country | United States | Maintains a clear leading position in research output and influence. |
| Emerging Contributor | China | Number of Chinese research participants has grown rapidly over the past decade. |
| Prolific Authors | Prof. Iadecola Costantino, Prof. Tarantini Stefano | Research of Prof. Tarantini Stefano is widely recognized in the field. |
| Dominant Research Terms | "cerebral blood flow," "neuronal activity," "neurovascular coupling" | Emphasizes the central role of brain function and hemodynamics. |
| Emerging Imaging Techniques | "fNIRS," "resting-state fMRI," "autoregulation" | Highlights growing impact of non-invasive neuroimaging. |
Cluster analysis of research themes has identified several focused areas, including functional connectivity, nitric oxide-mediated vascular regulation, cerebral autoregulation, Alzheimer's disease metabolism, and CO2-induced hemodynamic modulation [39]. The future of the field is expected to be driven by the integration of artificial intelligence, multi-omics analysis, and high-resolution imaging to further elucidate NVC mechanisms in health and disease [39].
Multimodal AI Frameworks for Data Integration and Analysis
Multimodal AI represents a pivotal evolution in artificial intelligence, allowing systems to process and merge data from diverse input formats such as text, images, audio, and video to grasp intricate contexts and provide precise insights [111] [112]. In biomedical research, these models can integrate imaging data with clinical notes, genomic information, and sensor data to form a comprehensive view of complex physiological processes like NVC [110].
Core Architectures and Fusion Techniques
A typical multimodal AI architecture consists of several key components that work in sequence to transform raw, multi-format data into actionable insights [112]:
- Encoders: These are modality-specific networks that transform raw data (e.g., images, text, audio) into machine-readable feature vectors or embeddings. Common encoders include Convolutional Neural Networks (CNNs) for images, transformer models for text, and models like Wav2Vec2 for audio [112].
- Fusion Mechanisms: This is the core of multimodal AI, where the extracted features from different modalities are combined. Fusion can occur at different stages and through various methods [112]:
- Early Fusion: Raw data from different modalities are combined before being fed into a single model.
- Intermediate Fusion: Latent representations (embeddings) of each modality are projected into a shared space and then fused.
- Late Fusion: Each modality is processed independently, and the final outputs or decisions are combined.
- Hybrid Fusion: A combination of the above strategies at different processing phases.
- Decoders: These networks process the fused feature vectors to generate the required output, which could be a classification, a prediction, or generated content (e.g., an image description). Decoders can be based on RNNs, CNNs, or Generative Adversarial Networks (GANs) [112].
Table 2: Comparison of AI Model Types in Biomedical Research
| Feature | Generative AI | Unimodal AI | Multimodal AI |
|---|---|---|---|
| Definition | AI designed to create new data or content. | AI that processes and understands a single type of data. | AI that integrates and processes multiple types of data. |
| Primary Use Cases | Automated text generation, image synthesis, content creation. | Language translation, image classification, speech recognition. | Autonomous driving, healthcare diagnostics, advanced surveillance. |
| Advantages | High creativity and realistic content generation. | High performance in specialized tasks, simplicity. | Rich insights and comprehensive understanding. |
| Training Data | Requires large, diverse datasets of the type it generates. | Needs datasets specific to the single data type it processes. | Utilizes large, diverse datasets covering multiple data types. |
| Challenges | Quality control, ethical considerations, computational demands. | Limited to a single modality, might miss context from other data types. | Integration complexity, higher computational requirements, data synchronization. |
| Examples | GPT-4, DALL-E, Stable Diffusion. | BERT (for text), ResNet (for images). | CLIP, GPT-4 Vision, Perceiver IO. |
Advanced Computational Frameworks for Biomedical Data
Two advanced frameworks are particularly suited for handling the complexities of multimodal biomedical data:
- Transformers: Originally developed for natural language processing, transformers employ a self-attention mechanism that allows them to weigh the importance of different parts of the input data. This is invaluable for understanding context in free-text clinical notes or genomic sequences. Their parallelized architecture allows for scalable computation, making them effective for integrating imaging, clinical, and genetic information to achieve high diagnostic accuracy, as demonstrated in Alzheimer's disease classification [110].
- Graph Neural Networks (GNNs): GNNs are designed to learn from non-Euclidean data structured as graphs. This is particularly relevant for biomedical data where relationships between entities—such as the connection between an anatomical region in a scan, a genetic marker, and a clinical symptom—are not grid-like. GNNs can model these complex interactions explicitly, providing a more natural and powerful representation than models that force data into a grid structure [110].
Experimental Protocols for Multimodal Neurovascular Investigation
Translating theoretical models into empirical discoveries requires rigorous experimental protocols. The following sections outline key methodologies for multimodal data acquisition and processing, with a specific focus on fNIRS, which is highly accessible but requires careful handling to avoid misinterpretation.
A Dynamic Causal Modelling (DCM) Framework for MEG/fMRI Fusion
The integration of electromagnetic (MEG/EEG) and hemodynamic (fMRI) data through Dynamic Causal Modelling (DCM) provides a powerful method for evaluating competing hypotheses about neurovascular coupling [109]. The procedure below outlines a Bayesian model comparison approach:
- fMRI Data Localization: The fMRI data are first used to identify regionally specific neuronal responses to a given paradigm. The coordinates of these activations serve as spatial priors for the subsequent model.
- DCM of Electrophysiological Data: The coordinates from Step 1 are used to specify a DCM for the MEG/EEG data collected during the same experimental paradigm. A neural mass model (e.g., a canonical microcircuit model) is inverted to explain the electrophysiological responses.
- Generation of Neuronal Drive Functions: The estimated parameters from the DCM are used to generate neuronal drive functions. These functions represent the pre- or post-synaptic activity for each experimental condition and each neuronal population in the model.
- Linking to Neurovascular Coupling: The neuronal drive functions form the input to a model of neurovascular coupling and the Balloon model for hemodynamics. The parameters of this combined model are estimated from the fMRI time-series data.
- Bayesian Model Comparison: The core of the procedure is to compare the evidence for different models of neurovascular coupling—for instance, asking whether the BOLD response is better explained by pre-synaptic or post-synaptic signals, or by inputs from specific neuronal populations [109].
The following workflow diagram visualizes this integrated methodological pipeline for combining MEG and fMRI data.
A Standardized fNIRS Data Processing Pipeline
fNIRS is susceptible to various noise sources, and suboptimal processing can significantly bias study results [113]. The following protocol details a robust processing pipeline to enhance signal quality and specificity to neurovascular coupling.
Channel Exclusion:
- Purpose: To identify and remove channels with poor signal quality due to poor optode-scalp contact, hair interference, or motion.
- Method: Automated criteria are preferred over subjective visual inspection. The PHOEBE (Placing Headgear Optodes Efficiently before Experimentation) method uses automatic detection of cardiac pulsation in the ~830-850 nm wavelength signal to determine adequate signal-to-noise ratio (SNR). The coefficient of variation (CV) method is another widely used automated alternative [113].
Motion Correction:
- Purpose: To identify and correct artifacts caused by subject movement, which can manifest as high-frequency spikes or baseline shifts in the data.
- Method: Wavelet-based filtering is highly effective, particularly for correcting high-frequency spikes. For data with significant baseline shifts, spline interpolation methods may be more effective. A visual inspection of the algorithm's performance on a subset of data is strongly recommended to ensure parameters are appropriately tuned [113].
Filtering and Denoising:
- Purpose: To remove physiological noise of non-neuronal origin (e.g., heartbeat, respiration, blood pressure waves) that can lead to false positives or negatives.
- Bandpass Filtering: A Butterworth bandpass filter is commonly applied to remove high-frequency (≥0.2 Hz) and very low-frequency (<0.01 Hz) noise [113].
- Denoising for Systemic Physiology: After bandpass filtering, additional methods are required to remove global systemic physiology. Common techniques include:
- Short Source-Detector (SSD) Regression: Uses a short-distance channel to measure and regress out extracerebral hemodynamics.
- Principal Component Analysis (PCA): A data-driven approach that removes signal components explaining the greatest spatial covariance across channels.
- Global Average (GloAvg) Removal: Removes the global average signal from all channels, though this method is debated due to concerns about introducing spurious correlations [113].
Statistical Analysis:
- Purpose: To model task-evoked hemodynamic responses and determine significant activation.
- Method: The General Linear Model (GLM) is widely used. It models the fNIRS time series as a linear combination of the expected hemodynamic response (convolved with the task timing) and several error terms. A multi-level analysis is then used to study differences between groups or conditions [113].
Computational Modeling of Neurovascular Coupling
Beyond AI-driven data fusion, mechanistic mathematical modeling provides a powerful tool for integrating knowledge across species and experimental modalities to form a unified quantitative understanding of NVC.
A Unified Cross-Species Quantitative Model
A landmark mathematical model integrates NVC data from mice, monkeys, and humans to preserve mechanistic insights across species [4] [3]. This model connects three critical layers:
- Cellular Signaling Model: Derived from optogenetics and microscopy data in mice, this layer describes cell-specific contributions. For example, it quantifies how NO-interneurons cause the initial rapid dilation, pyramidal neurons drive the main dilation, and NPY-interneurons generate the post-peak undershoot [3].
- Windkessel Hemodynamic Model: This component models blood flow, pressure, and volume in arterioles, capillaries, and venules. It translates the vasoactive signals from the cellular model into changes in Cerebral Blood Flow (CBF) and Cerebral Blood Volume (CBV) [3].
- BOLD Signal Model: The final layer uses the Griffeth and Buxton model to integrate the outputs from the hemodynamic model with changes in the Cerebral Metabolic Rate of Oxygen (CMRO2) to generate a predicted BOLD signal [3].
The key innovation of this approach is its "preservation of qualitative insights". Mechanistic behaviors discovered in highly controlled animal studies (e.g., the role of specific interneurons) are translated and preserved as fixed qualitative constraints when the model is fitted to human data, which might include additional measurements of blood flow and volume in arterioles and venules [3]. This allows for a more biologically grounded and interpretable analysis of human neuroimaging data.
The following table details key reagents, computational tools, and data types essential for advanced research at the intersection of multimodal AI and neurovascular coupling.
Table 3: Research Reagent Solutions for Multimodal Neurovascular Research
| Resource Category | Specific Examples | Function and Application |
|---|---|---|
| Computational Modeling Tools | Dynamic Causal Modelling (DCM) in SPM, HOMER2 (for fNIRS) | Provides frameworks for Bayesian model comparison of neurovascular hypotheses and standardized processing of fNIRS data [109] [113]. |
| Multimodal AI Models & Frameworks | CLIP, DALL-E, LLaVA, CogVLM, Transformer Models, Graph Neural Networks (GNNs) | Enable tasks like visual question-answering, image-text fusion, and learning from complex, non-Euclidean biomedical data relationships [112] [110]. |
| Key Experimental Data Types | Optogenetic recordings, Local Field Potential (LFP), BOLD-fMRI, fNIRS (HbO/HbR), Arteriolar Diameter | Provides complementary information for building and validating cross-species quantitative models of NVC [4] [3]. |
| Cell-Type Specific Insights | NO-interneurons, NPY-interneurons, Pyramidal Neurons, Astrocytic Signaling Pathways | Define specific temporal components (rapid dilation, undershoot) and chemical pathways in the neurovascular response [4] [3]. |
The integration of multimodal AI and advanced computational models is fundamentally reshaping the landscape of neurovascular research. This synergy moves the field beyond simplistic correlations, enabling a mechanistically resolved understanding of the link between neural activity and hemodynamic signals. For researchers and drug development professionals, this paradigm shift offers a clear path toward more reliable biomarkers and therapeutic targets. The application of transformer-based networks and GNNs to fused datasets—encompassing fMRI, fNIRS, MEG, and clinical metadata—will unlock unprecedented precision in characterizing brain function and dysfunction. As these multimodal AI frameworks continue to evolve, they will not only enhance the interpretability of functional neuroimaging but also pave the way for personalized medicine in neurology and psychiatry, ultimately leading to more effective diagnostic and therapeutic strategies for complex brain disorders.
Conclusion
Neurovascular coupling serves as the critical, though indirect, bridge between neural activity and the non-invasive measurements provided by fMRI and fNIRS. A deep understanding of its mechanisms is paramount for accurately interpreting functional imaging data in both health and disease. The future of NVC research lies in the sophisticated integration of multi-modal techniques, such as simultaneous EEG-fNIRS, to directly validate coupling dynamics. Furthermore, the application of artificial intelligence and multi-omics approaches promises to unravel the complex pathophysiology of NVC dysfunction. For biomedical and clinical research, this translates into the development of NVC as a sensitive, early biomarker for diagnosing neurodegenerative and cerebrovascular diseases and for evaluating the efficacy of novel therapeutics aimed at rescuing vascular and metabolic function in the brain.
References
- 1. Investigating Human Neurovascular Coupling Using ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4683196/]
- 2. Neurovascular Coupling - an overview [https://www.sciencedirect.com/topics/neuroscience/neurovascular-coupling]
- 3. A quantitative model for human neurovascular coupling with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9821752/]
- 4. A quantitative model for human neurovascular coupling with ... [https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1010818]
- 5. Cells of the Blood-brain Barrier - PubMed Central - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC9987262/]
- 6. Neurovascular unit [https://en.wikipedia.org/wiki/Neurovascular_unit]
- 7. Neurovascular Coupling: Scientometric Analysis of 30 Years ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12606022/]
- 8. Neurovascular Coupling by Functional Near Infra-Red ... [https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2020.00042/full]
- 9. Neurovascular coupling abnormalities in first-episode drug ... [https://bmcpsychiatry.biomedcentral.com/articles/10.1186/s12888-025-07503-x]
- 10. Fundamental Neurovascular Components for the ... [https://www.heraldopenaccess.us/openaccess/fundamental-neurovascular-components-for-the-development-of-complex-and-dynamic-in-vitro-brain-equivalent-models]
- 11. From Mechanisms to Medicine: Neurovascular Coupling in the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11719775/]
- 12. Investigating Human Neurovascular Coupling Using ... [https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2015.00467/full]
- 13. Neurovascular coupling mechanisms in health and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9337814/]
- 14. Contribution of astrocytes to neurovascular coupling in the ... [https://www.sciencedirect.com/science/article/pii/S1880654624002075]
- 15. What Are the Roles of Pericytes in the Neurovascular Unit and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10186232/]
- 16. Pericytes and Neurovascular Function in the Healthy and ... [https://www.frontiersin.org/journals/cellular-neuroscience/articles/10.3389/fncel.2019.00282/full]
- 17. Nitric Oxide Pathways in Neurovascular Coupling Under ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8569710/]
- 18. An EEG-fNIRS neurovascular coupling analysis method to ... [https://www.sciencedirect.com/science/article/abs/pii/S001048252300433X]
- 19. Redox and Nitric Oxide-Mediated Regulation of Sensory ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4323017/]
- 20. Flow-mediated vasodilation through mechanosensitive G ... [https://www.sciencedirect.com/science/article/abs/pii/S1050173821000013]
- 21. Pharmacologically induced impairment of neurovascular ... [https://pubmed.ncbi.nlm.nih.gov/29243191/]
- 22. Inducing a Functional-Pharmacological Coupling in the ... [https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2020.557874/full]
- 23. Neurovascular coupling in humans: Physiology ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4821024/]
- 24. The confound of hemodynamic response function ... [https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2023.934138/full]
- 25. The Hemo-Neural Hypothesis: On The Role of Blood Flow in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3655718/]
- 26. The role of fMRI in drug development - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC5931333/]
- 27. Neurovascular Coupling in Development and Disease [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2021.702832/full]
- 28. Towards whole brain mapping of the haemodynamic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11994648/]
- 29. Optimal hemodynamic response model for functional near- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4468613/]
- 30. Wearable fNIRS platform for dense sampling and precision ... [https://www.nature.com/articles/s41746-025-01690-3]
- 31. Evaluation of hemodynamic response function during ... [https://www.sciencedirect.com/science/article/pii/S2772528621000042]
- 32. Neurovascular coupling during auditory stimulation [https://pmc.ncbi.nlm.nih.gov/articles/PMC10517045/]
- 33. Evaluation of fNIRS signal components elicited by cognitive ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8648757/]
- 34. Traces of EEG-fMRI coupling reveals neurovascular ... [https://www.nature.com/articles/s41598-024-51694-4]
- 35. Physiologically informed dynamic causal modeling of fMRI ... [https://www.sciencedirect.com/science/article/pii/S1053811915006989]
- 36. Applications and advances of combined fMRI-fNIRs ... [https://www.frontiersin.org/journals/neurology/articles/10.3389/fneur.2025.1542075/full]
- 37. A tight relationship between BOLD fMRI activation ... [https://pubmed.ncbi.nlm.nih.gov/40793082/]
- 38. A tight relationship between BOLD fMRI activation/ ... [https://elifesciences.org/articles/104779]
- 39. Scientometric Analysis of 30 Years Research (1996-2025) [https://pubmed.ncbi.nlm.nih.gov/41220183/]
- 40. Inhibitory neurons dominate BOLD-fMRI responses [https://www.sciencedirect.com/science/article/pii/S0010482525013666]
- 41. The physiological component of the BOLD signal: Impact ... [https://www.vanderbilt.edu/valiant/2025/08/25/the-physiological-component-of-the-bold-signal-impact-of-age-and-heart-rate-variability-biofeedback-training/]
- 42. Comparison of Brain Oxygen Metabolic Parameters ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12337088/]
- 43. Apparent Diffusion Coefficient fMRI shines light on white ... [https://www.nature.com/articles/s42003-025-07889-0]
- 44. Functional Magnetic Resonance Imaging and Functional Near ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5563305/]
- 45. Comparison of fNIRS with other neuroimaging modalities [https://www.artinis.com/blogpost-all/comparison-fnirs-vs-fmri]
- 46. Applications of Functional Near-Infrared Spectroscopy ... [https://www.mdpi.com/2077-0383/14/15/5197]
- 47. Phasor representation of oxy- and deoxyhemoglobin ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2941517/]
- 48. Coupling of Oxy- and Deoxyhemoglobin concentrations ... [https://www.nature.com/articles/s41598-017-15770-2]
- 49. fNIRS - fMRI | Concurrent NIRS MRI Recording ... [https://nirx.net/fnirs-fmri]
- 50. The Effects of Sensory Manipulations on Motor Behavior [https://pmc.ncbi.nlm.nih.gov/articles/PMC6124483/]
- 51. Cognitive and Motor Learning in Internally-Guided ... [https://www.frontiersin.org/journals/psychology/articles/10.3389/fpsyg.2021.604323/full]
- 52. Impaired neurovascular coupling in metabolic syndrome [https://pubmed.ncbi.nlm.nih.gov/40387835/]
- 53. Neurovascular Coupling by Functional Near Infra-Red ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7033387/]
- 54. Protocol for activity flow mapping of neurocognitive ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8808261/]
- 55. Data-guided neuroimaging and visualization [https://apertureneuro.org/article/140427-data-guided-neuroimaging-and-visualization-from-functional-decomposition-to-dynamic-fusion]
- 56. Implications of neurovascular uncoupling in functional ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5669348/]
- 57. Functional Near-Infrared Spectroscopy and Its Clinical ... [https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2020.00724/full]
- 58. Functional Near-Infrared Spectrometry as a Useful ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10779649/]
- 59. The present and future use of functional near‐infrared ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6367070/]
- 60. A Mini-Review on Functional Near-Infrared Spectroscopy ... [https://www.mdpi.com/2304-6732/6/3/87]
- 61. Functional near-infrared spectroscopy [https://en.wikipedia.org/wiki/Functional_near-infrared_spectroscopy]
- 62. A multimodal EEG-fNIRS-TCD investigation [https://www.sciencedirect.com/science/article/pii/S1053811924004075]
- 63. The effects of protocol factors and participant ... [https://www.sciencedirect.com/science/article/pii/S2666956025000443]
- 64. Systemic physiology augmented functional near-infrared ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9272976/]
- 65. Modelling confounding effects from extracerebral ... [https://www.sciencedirect.com/science/article/pii/S1053811916304505]
- 66. Adaptive filtering of physiological noises in fNIRS data [https://biomedical-engineering-online.biomedcentral.com/articles/10.1186/s12938-018-0613-2]
- 67. Effects of Systemic Physiology on Mapping Resting-State ... [https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2022.803297/full]
- 68. Motion artifacts in functional near-infrared spectroscopy [https://pmc.ncbi.nlm.nih.gov/articles/PMC3762942/]
- 69. fNIRS reproducibility varies with data quality, analysis ... [https://www.nature.com/articles/s42003-025-08412-1]
- 70. Functional near-infrared spectroscopy for the assessment ... [https://www.frontiersin.org/journals/neurology/articles/10.3389/fneur.2025.1524806/full]
- 71. A Novel Optimisation Framework for fNIRS [https://pubmed.ncbi.nlm.nih.gov/40857195/]
- 72. Development of Wearable Wireless Multichannel f - NIRS System to... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12114292/]
- 73. Functional connectivity in whole-brain and network analysis ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-025-07181-z]
- 74. Frontiers | Impact of hearing loss on brain signal variability in older... [https://www.frontiersin.org/journals/aging-neuroscience/articles/10.3389/fnagi.2025.1498666/full]
- 75. MetaNIRS: A General Decoding Framework for fNIRS ... [https://www.sciencedirect.com/science/article/abs/pii/S0893608025007531]
- 76. The hemodynamic inverse problem: Making inferences ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC151314/]
- 77. Neurovascular coupling alteration in drug-naïve ... [https://www.sciencedirect.com/science/article/pii/S0969996124000056]
- 78. Modeling the Hemodynamic Response Function in fMRI [https://pmc.ncbi.nlm.nih.gov/articles/PMC3318970/]
- 79. Advancements in Numerical Methods for Forward and ... [https://www.mdpi.com/2075-1680/12/4/326]
- 80. Fusion of fNIRS and fMRI data: identifying when and where ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3797402/]
- 81. Brain endothelial gap junction coupling enables rapid ... [https://www.sciencedirect.com/science/article/pii/S0092867425007329]
- 82. The hemodynamic model solving algorithm by using fMRI ... [https://www.sciencedirect.com/science/article/pii/S2772528622000541]
- 83. Visual task complexity and eye movement patterns ... [https://www.sciencedirect.com/science/article/abs/pii/S0031938420305126]
- 84. Data Processing in Functional Near-Infrared Spectroscopy ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8151801/]
- 85. During Auditory... | Neurovascular Square Coupling Research [https://www.researchsquare.com/article/rs-2827122/v1]
- 86. fMRI-based validation of continuous-wave fNIRS ... [https://www.nature.com/articles/s41598-022-06519-7]
- 87. Brain–Computer Interface-Robot Training Enhances Upper ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9024364/]
- 88. Neurovascular coupling in eye-open-eye-close task and ... [https://www.sciencedirect.com/science/article/pii/S1053811924000302]
- 89. Neuromodulation of Cerebral Blood Flow: A Physiological ... [https://www.mdpi.com/2306-5354/12/5/442]
- 90. Applications and advances of combined fMRI-fNIRs ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11958174/]
- 91. Traces of EEG-fMRI coupling reveals neurovascular dynamics ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10794702/]
- 92. Physiological influences on neurovascular coupling [https://pmc.ncbi.nlm.nih.gov/articles/PMC11689421/]
- 93. Machine learning: assessing neurovascular signals in the ... [https://www.nature.com/articles/s41598-019-54316-6]
- 94. Low frequency oscillations reflect neurovascular coupling ... [https://www.nature.com/articles/s41598-024-61819-4]
- 95. Evaluation on the application of transcranial Doppler (TCD) ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7557013/]
- 96. Applications of near-infrared spectroscopy in neurocritical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10311235/]
- 97. A Systematic Review of Cerebral Functional Near-Infrared ... [https://www.mdpi.com/2075-4418/10/8/581]
- 98. Concurrent fNIRS and EEG for Brain Function Investigation [https://pmc.ncbi.nlm.nih.gov/articles/PMC9371171/]
- 99. fNIRS vs EEG: Comparing Non-Invasive Brain Imaging ... [https://imotions.com/blog/learning/research-fundamentals/fnirs-vs-eeg/?srsltid=AfmBOopO4KQxNxMDRFl8_iDvwvapiFa5Iyu73DWGDZ9uQvWmIzJxdYxq]
- 100. A Cross-Disciplinary Review of the fNIRS-EEG Dual-Modality ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11506513/]
- 101. High-density multidistance fNIRS enhances detection of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12412631/]
- 102. Optimizing spatial specificity and signal quality in fNIRS [https://pmc.ncbi.nlm.nih.gov/articles/PMC11188482/]
- 103. Limitations of temporal resolution in functional MRI - PubMed [https://pubmed.ncbi.nlm.nih.gov/9094089/]
- 104. Hemodynamic timing in resting-state and breathing-task ... [https://www.sciencedirect.com/science/article/pii/S1053811923002665]
- 105. Surface-based integration approach for fNIRS-fMRI ... [https://www.sciencedirect.com/science/article/abs/pii/S0165027023001711]
- 106. common mechanisms and strategies for new treatments [https://molecularneurodegeneration.biomedcentral.com/articles/10.1186/s13024-022-00524-0]
- 107. Neurovascular coupling, functional connectivity, and ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11350141/]
- 108. Near-infrared spectroscopy of the cerebellum in motor ... [https://www.nature.com/articles/s41598-025-04678-x]
- 109. Comparing dynamic causal models of neurovascular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7322559/]
- 110. The future of multimodal artificial intelligence models for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12239537/]
- 111. Top 10 Innovative Multimodal AI Applications and Use Cases [https://appinventiv.com/blog/multimodal-ai-applications/]
- 112. Top 10 Multimodal Models [https://encord.com/blog/top-multimodal-models/]
- 113. Automated Processing of fNIRS Data—A Visual Guide to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6428450/]