Synergizing EEG and fNIRS: A Comprehensive Guide to Hybrid Brain-Computer Interface Systems for Biomedical Research
This article provides a detailed exploration of simultaneous EEG-fNIRS setups for brain-computer interface (BCI) applications, tailored for researchers, scientists, and drug development professionals.
Synergizing EEG and fNIRS: A Comprehensive Guide to Hybrid Brain-Computer Interface Systems for Biomedical Research
Abstract
This article provides a detailed exploration of simultaneous EEG-fNIRS setups for brain-computer interface (BCI) applications, tailored for researchers, scientists, and drug development professionals. It covers the foundational principles of both modalities, highlighting their complementary nature—EEG's millisecond-scale temporal resolution for electrical activity and fNIRS's superior spatial resolution for hemodynamic responses. The content delves into practical methodological aspects of system integration, data acquisition, and signal processing, alongside advanced fusion strategies and analysis techniques. Furthermore, it addresses common troubleshooting challenges, optimization methods for enhanced performance, and a comparative validation of the hybrid system's efficacy against unimodal approaches through case studies and performance metrics. The article concludes by synthesizing key takeaways and outlining future directions for the technology in clinical and biomedical research applications.
Understanding the Core Principles: Why EEG and fNIRS are a Powerful Combination for BCI
In brain-computer interface (BCI) research, the quest for a more comprehensive understanding of brain activity has driven the adoption of multimodal neuroimaging. Electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS) have emerged as a particularly powerful combination, integrating distinct yet complementary information about neuroelectrical and neurovascular activities [1] [2]. EEG measures the brain's rapid electrical activity resulting from the summation of post-synaptic potentials of pyramidal neurons, providing millisecond-level temporal resolution but limited spatial precision [3] [4]. In contrast, fNIRS utilizes near-infrared light to monitor hemodynamic responses linked to neural metabolism, offering superior spatial localization but slower temporal resolution [1] [5]. This inherent complementarity enables researchers to capture a more complete picture of brain dynamics, making simultaneous EEG-fNIRS particularly valuable for BCI systems aimed at decoding diverse cognitive states and intents [6] [7].
Fundamental Signal Origins and Complementarity
Electroencephalography (EEG) Signal Origins
EEG captures electrical potentials generated primarily by the synchronized postsynaptic activity of cortical pyramidal neurons. When these neurons fire in synchrony, the resulting current flows create electrical fields measurable at the scalp surface [3]. The signal is characterized by its excellent temporal resolution (milliseconds) but suffers from limited spatial resolution due to the blurring effects of the skull, meninges, and other tissues between the cortex and electrodes [2] [4]. This fundamental physical property makes EEG ideal for tracking rapid neural dynamics but challenges precise localization of neural sources.
Functional Near-Infrared Spectroscopy (fNIRS) Signal Origins
fNIRS relies on neurovascular coupling, the tight relationship between neural activity and subsequent hemodynamic changes. It employs near-infrared light (650-950 nm wavelengths) to measure concentration changes in oxygenated hemoglobin (HbO) and deoxygenated hemoglobin (HbR) in the cerebral cortex [1] [5]. Active brain regions experience increased oxygen demand, triggering a compensatory hemodynamic response that alters the relative concentrations of HbO and HbR – a phenomenon fNIRS detects through differential light absorption properties of these chromophores [1] [8]. This optical measurement provides good spatial resolution but is inherently limited by the slow nature of the hemodynamic response (typically peaking 4-6 seconds after neural activation) [3].
Complementary Characteristics for BCI
The orthogonal nature of EEG and fNIRS signals creates powerful synergies for BCI applications. Their complementary properties span temporal and spatial domains, sensitivity to artifacts, and the types of brain activity they best capture [2]. The following table summarizes these complementary characteristics:
Table 1: Complementary Characteristics of EEG and fNIRS for BCI Applications
| Feature | EEG | fNIRS |
|---|---|---|
| Temporal Resolution | Millisecond precision [3] | Slower (0.1-0.5 Hz), peaks 4-6s post-stimulus [3] |
| Spatial Resolution | Limited (several cm) due to volume conduction [2] [3] | Better (<1 cm) for cortical mapping [3] |
| Signal Origin | Electrical activity (post-synaptic potentials) [3] | Hemodynamic response (HbO/HbR concentration changes) [1] |
| Artifact Sensitivity | Sensitive to electrical noise, muscle artifacts, eye movements [4] | Sensitive to scalp blood flow, motion artifacts, ambient light [1] |
| Measured Process | Direct neural electrical activity [9] | Metabolic demand following neural activity [9] |
| Ideal BCI Tasks | Rapid changes (P300, SSVEP, motor imagery onset) [5] [9] | Sustained cognitive states (mental arithmetic, workload, vigilance) [5] [9] |
The signaling pathways illustrating the relationship between neural activity and the measurable signals for each modality can be visualized as follows:
Diagram 1: Signal Origin Pathways
Quantitative Performance in BCI Applications
Research has consistently demonstrated that combining EEG and fNIRS yields superior BCI performance compared to either modality alone. The integration enhances classification accuracy across various paradigms, particularly for motor imagery and cognitive tasks.
Table 2: BCI Classification Performance of Unimodal vs. Multimodal Approaches
| Modality | Task | Key Features/Methods | Reported Performance | Citation |
|---|---|---|---|---|
| EEG-only | Motor Imagery (MI) | Event-related desynchronization | Lower accuracy compared to hybrid | [4] |
| fNIRS-only | Mental Arithmetic (MA) | HbO/HbR mean, slope, variance | Lower accuracy compared to hybrid | [4] |
| EEG-fNIRS Hybrid | MI & MA | Multi-domain features + multi-level progressive learning | 96.74% (MI), 98.42% (MA) accuracy | [6] [4] |
| EEG-fNIRS Hybrid | MI | Non-linear features + ensemble learning | 95.48% accuracy, 97.67% F1-score | [7] |
| EEG-fNIRS Hybrid | Mental Stress | Decision-level fusion (SVM probability combining) | +7.76% vs. EEG, +10.57% vs. fNIRS | [4] |
Experimental Protocols for Simultaneous EEG-fNIRS
Protocol 1: Motor Imagery and Mental Arithmetic Paradigm
This protocol adapts well-established tasks that elicit robust responses in both modalities, suitable for evaluating hybrid BCI performance [6] [4].
Materials and Setup:
- Simultaneous EEG-fNIRS system with synchronized data acquisition
- 64-channel EEG cap integrated with fNIRS optodes
- fNIRS source-detector pairs over motor and prefrontal cortices
- Visual stimulus presentation system
- Trigger synchronization interface (LSL or hardware triggers)
Procedure:
- Participant Preparation: Fit integrated EEG-fNIRS cap ensuring proper electrode impedance (<10 kΩ for EEG) and optical contact quality for fNIRS [3] [9].
- Baseline Recording: Acquire 5 minutes of resting-state data with eyes open for later signal normalization.
- Task Block Structure: Implement alternating task and rest blocks:
- Trial Marking: Implement event markers for both brief EEG events (stimulus onset) and extended fNIRS blocks (task period start/end) [3].
- Data Acquisition: Record continuous EEG (≥500 Hz sampling) and fNIRS (10-50 Hz sampling) with synchronized triggers.
Data Analysis:
- EEG Processing: Bandpass filtering (0.5-40 Hz), artifact removal (ICA), time-locked epoch extraction, and feature extraction (power spectral densities, ERD/ERS) [4].
- fNIRS Processing: Conversion of optical density to HbO/HbR concentrations, bandpass filtering (0.01-0.2 Hz), motion artifact correction, and feature extraction (mean, slope, variance) [6] [5].
- Multimodal Fusion: Apply feature-level fusion (concatenating EEG and fNIRS features) or decision-level fusion (combining classifier outputs) [6] [7].
The experimental workflow for a typical simultaneous recording session proceeds through distinct phases:
Diagram 2: Experimental Workflow
Protocol 2: Resting-State Functional Connectivity with Vigilance Monitoring
This protocol examines the relationship between electrophysiological and hemodynamic signals during resting states, particularly valuable for clinical populations.
Materials and Setup:
- High-density EEG-fNIRS systems (whole-head coverage)
- Polysomnography-capable amplifier for EEG vigilance monitoring
- Multiple short-distance fNIRS channels for superficial signal regression [10]
- Comfortable reclining chair with support for various body positions
Procedure:
- System Setup: Configure high-density montage (≥64 EEG channels, ≥64 fNIRS channels) with emphasis on frontal and default mode network regions.
- Position Variation: Acquire data in multiple body positions (supine, sitting, standing) to assess posture effects on global signals [10].
- Extended Recording: Collect 45 minutes of resting-state data with simultaneous EEG-fNIRS for vigilance fluctuation analysis.
- Vigilance Assessment: Continuously monitor EEG for vigilance shifts using standardized rating scales (e.g., VIGALL).
- Control Task: Include auditory oddball paradigm to assess evoked responses across body positions.
Data Analysis:
- Global Signal Analysis: Calculate fNIRS global signal as average across all channels, examine frequency content (0.01-0.1 Hz) [10].
- Vigilance Correlation: Compute correlation between EEG vigilance measures and fNIRS global signal amplitude.
- Functional Connectivity: Assess RSFC using correlation-based approaches between brain regions.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful simultaneous EEG-fNIRS experimentation requires careful selection of equipment and materials that address the unique challenges of multimodal integration.
Table 3: Essential Materials for Simultaneous EEG-fNIRS Research
| Item | Specification/Type | Function/Purpose | Implementation Notes |
|---|---|---|---|
| Integrated Cap System | Customizable cap with 128-160 slits, black fabric | Hosts both EEG electrodes and fNIRS optodes, prevents light leakage | actiCAP with 128 slits recommended; dark fabric reduces optical reflection [3] |
| EEG Electrodes | Active electrode systems (e.g., g.SCARABEO) | Measures electrical potentials with high signal quality | Active electrodes reduce preparation time to ~10 minutes for 32 channels [9] |
| fNIRS Optodes | Laser diode or LED sources with sensitive detectors | Measures hemodynamic responses via light absorption | Source-detector distance of 20-30 mm standard; multiple wavelengths (e.g., 760, 850 nm) [1] [9] |
| Synchronization Interface | LSL protocol or shared hardware triggers | Ensures temporal alignment of EEG and fNIRS data streams | Critical for event-related analysis; LSL enables software synchronization [3] |
| Amplifier System | Hybrid EEG-fNIRS amplifiers (e.g., g.Nautilus with g.SENSOR fNIRS) | Simultaneously acquires both signal types with minimal interference | Integrated systems simplify setup and improve synchronization precision [9] |
| Montage Design Software | MATLAB-based tools (e.g., ArrayDesigner) | Plans optimal placement of competing sensors | Determines trade-offs between EEG and fNIRS coverage in target regions [3] |
Integration Methodologies and Fusion Approaches
The successful integration of EEG and fNIRS data occurs at multiple levels, each with distinct advantages for BCI applications. The three primary fusion strategies include:
Data-Level Fusion: Direct combination of raw or minimally processed signals, though this approach is computationally intensive and less commonly used due to the fundamentally different nature of EEG and fNIRS signals [4].
Feature-Level Fusion: This dominant approach involves extracting relevant features from each modality then combining them into a unified feature vector for classification. Methods range from simple concatenation to advanced techniques like multi-domain feature extraction with optimization algorithms [6]. Recent advances include multi-level progressive learning frameworks that achieve >96% accuracy in both motor imagery and mental arithmetic tasks [6] [4].
Decision-Level Fusion: Separate classification of each modality followed by combination of decisions through voting schemes, weighted averaging, or meta-classifiers. This approach provides robustness against modality-specific artifacts and has demonstrated significant improvements (+7-10%) over unimodal approaches in mental stress detection [4].
The relationship between these fusion approaches and their respective advantages can guide selection based on specific BCI requirements:
Diagram 3: Multimodal Fusion Approaches
EEG and fNIRS provide fundamentally distinct yet highly complementary windows into brain function, with electrical and hemodynamic signals offering temporally and spatially orthogonal information. Their successful integration in simultaneous setups requires careful attention to experimental design, hardware integration, and data fusion methodologies. The protocols and frameworks presented here provide a foundation for leveraging this powerful multimodal approach in BCI research, enabling more robust and comprehensive decoding of brain states and intents. As hardware integration continues to advance and analysis techniques become increasingly sophisticated, simultaneous EEG-fNIRS is poised to become an increasingly indispensable tool in the neuroscience and neuroengineering toolkit.
The development of brain-computer interfaces (BCIs) necessitates neuroimaging techniques that can accurately decode neural activity with high precision in both time and space. Electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS) have emerged as leading non-invasive modalities, each with distinct resolution profiles that present researchers with a fundamental trade-off. EEG measures the brain's electrical activity via electrodes placed on the scalp, offering millisecond-level temporal resolution but limited spatial accuracy due to the dispersion of electrical signals as they pass through the skull and scalp [11]. In contrast, fNIRS monitors cerebral hemodynamic responses by measuring changes in oxygenated and deoxygenated hemoglobin using near-infrared light, providing superior spatial resolution for surface cortical areas but constrained by a slower temporal response on the scale of seconds [11] [12].
This application note examines the temporal versus spatial resolution trade-offs between EEG and fNIRS within the context of simultaneous setup for BCI research. The complementary nature of these modalities enables a hybrid approach that mitigates their individual limitations through strategic integration [13]. We provide a comprehensive analysis of their comparative strengths, methodological protocols for simultaneous implementation, and visualization of integrated signaling pathways to guide researchers in optimizing BCI system design.
Technical Comparison of EEG and fNIRS
Fundamental Principles and Signal Characteristics
Electroencephalography (EEG) captures postsynaptic potentials generated primarily by pyramidal cells in the cerebral cortex. When tens of thousands of these neurons fire synchronously, with dendritic trunks oriented parallel to each other and perpendicular to the cortical surface, their electrical signals summate sufficiently to be detected by scalp electrodes [12]. These signals represent large-scale neural oscillatory activity divided into characteristic frequency bands: theta (4-7 Hz), alpha (8-14 Hz), beta (15-25 Hz), and gamma (>25 Hz) [12].
Functional Near-Infrared Spectroscopy (fNIRS) employs near-infrared light (600-1000 nm wavelength) to measure hemodynamic responses coupled with neural activity. Light at different wavelengths is introduced into the scalp via optical sources, and the attenuated light that diffusely reflects back to detectors is used to compute concentration changes in oxygenated hemoglobin (HbO) and deoxygenated hemoglobin (HbR) based on the Modified Beer-Lambert Law [12] [5]. These hemodynamic changes serve as indirect markers of brain activity through the mechanism of neurovascular coupling [12].
Comparative Analysis of Resolution Characteristics
Table 1: Quantitative Comparison of EEG and fNIRS Resolution Profiles
| Parameter | EEG | fNIRS |
|---|---|---|
| Temporal Resolution | Milliseconds [11] | Seconds (typically 2-6 second delay) [11] [14] |
| Spatial Resolution | Centimeter-level [11] | Moderate (better than EEG, limited to cortex) [11] |
| Depth of Measurement | Cortical surface [11] | Outer cortex (approximately 1-2.5 cm deep) [11] |
| Signal Source | Postsynaptic potentials in cortical neurons [11] | Hemodynamic response (blood oxygenation changes) [11] |
| Neurovascular Coupling | Direct neural electrical activity [12] | Indirect metabolic-hemodynamic response [12] |
| Movement Artifact Sensitivity | High susceptibility [11] [15] | Relatively robust tolerance [11] [15] |
The fundamental trade-off between temporal and spatial resolution emerges from the different physiological phenomena each modality captures. EEG provides a direct view of neural dynamics with exceptional temporal fidelity, making it ideal for tracking rapid cognitive processes like stimulus perception and decision onset [11]. However, its spatial resolution is limited due to the blurring effect of the skull and scalp on electrical fields [11] [12].
Conversely, fNIRS offers superior spatial localization for surface cortical areas, particularly the prefrontal region, but is constrained by the inherent delay of the hemodynamic response, which typically peaks 4-6 seconds after neural activation [11] [15]. This temporal lag presents challenges for real-time BCI applications requiring immediate feedback [15].
Neurophysiological Basis for Integration
The Neurovascular Coupling Mechanism
The theoretical foundation for integrating EEG and fNIRS lies in the physiological phenomenon of neurovascular coupling - the intimate relationship between neural electrical activity and subsequent hemodynamic responses [12]. When neurons within a specific brain region activate, they trigger a complex cascade of metabolic and vascular events that increase local cerebral blood flow to deliver oxygen and nutrients, resulting in measurable fluctuations in hemoglobin concentrations [12].
This coupling forms the basis for connecting EEG's direct measurement of electrical activity with fNIRS's indirect measurement of hemodynamic changes, providing complementary information about the same underlying neural events [12]. Importantly, impairments in neurovascular coupling have been associated with various neurological conditions, including Alzheimer's disease and stroke, making simultaneous measurement valuable for both basic research and clinical applications [12].
Signaling Pathway in Simultaneous EEG-fNIRS
The following diagram illustrates the integrated signaling pathway from neural activity to measured signals in simultaneous EEG-fNIRS experimentation:
Signaling Pathway from Neural Activity to Measured Signals
This pathway visualization illustrates the parallel processes by which underlying neural activity generates measurable EEG and fNIRS signals. The direct electrical pathway (blue) enables millisecond temporal resolution, while the indirect hemodynamic pathway (red) provides superior spatial localization at the cost of slower response time [12].
Experimental Protocols for Simultaneous EEG-fNIRS
System Setup and Sensor Placement
Equipment Configuration:
- Utilize integrated EEG-fNIRS systems or synchronized separate systems with trigger synchronization [11]
- For EEG: Employ systems with 16+ channels, sampling rate ≥500 Hz (e.g., BrainAmp DC EEG system) [15]
- For fNIRS: Implement continuous-wave systems with dual-wavelength sources (e.g., 760 nm and 850 nm) and photodetectors [12] [15]
- Ensure emitter-detector distance of 30 mm for optimal sensitivity to cerebral tissues [16]
Sensor Placement Protocol:
- Use the international 10-20 or 10-5 system for consistent positioning [11] [16]
- For motor imagery tasks: Focus on C3, C4 positions with coverage of prefrontal cortex [15]
- For cognitive tasks: Emphasize prefrontal cortex coverage [5]
- Implement high-density EEG caps with pre-defined fNIRS-compatible openings to avoid physical interference [11]
- Use optode holders that avoid electrode contact points to prevent signal contamination [11]
Data Acquisition and Preprocessing Workflow
Table 2: Research Reagent Solutions for EEG-fNIRS Experimentation
| Category | Specific Solution/Equipment | Function/Purpose |
|---|---|---|
| fNIRS Hardware | NIRScout System (NIRx) [16] | Continuous-wave fNIRS data acquisition |
| EEG Hardware | BrainAmp DC EEG System (Brain Products) [15] | High-quality EEG signal acquisition |
| Sensor Integration | Integrated EEG-fNIRS Caps [11] | Compatible sensor placement minimizing interference |
| Synchronization | TTL Pulse Systems/Parallel Port Triggers [11] | Temporal alignment of EEG and fNIRS data streams |
| fNIRS Processing | Modified Beer-Lambert Law [12] [5] | Conversion of optical density to hemoglobin concentration changes |
| Artifact Removal | Motion Correction Algorithms [11] | Reduction of movement artifacts in both modalities |
| Advanced Analysis | Joint Independent Component Analysis (jICA) [11] | Multimodal data fusion and feature extraction |
The experimental workflow for simultaneous acquisition involves:
Simultaneous EEG-fNIRS Experimental Workflow
Data Acquisition Protocol:
- Record EEG signals at minimum 250-500 Hz sampling rate [15]
- Record fNIRS signals at 10-12.5 Hz sampling rate [14] [16]
- Implement synchronized task paradigms with randomized trial sequences
- For motor execution/imagery: Use block designs with 20s rest, 5s task periods [15]
- Include sufficient inter-trial intervals to allow hemodynamic response return to baseline
Preprocessing Guidelines:
- EEG Processing: Apply bandpass filtering (0.5-40 Hz), remove ocular and motion artifacts using independent component analysis (ICA) [11]
- fNIRS Processing: Convert raw intensity signals to optical density, then to HbO/HbR concentrations using Modified Beer-Lambert Law [12] [5]. Apply bandpass filtering (0.01-0.2 Hz) to remove physiological noise [5]
- Temporal Alignment: Address inherent fNIRS hemodynamic delay (typically 2-6 seconds) through temporal correction algorithms [14]
Application in Brain-Computer Interface Research
Task-Specific Implementation Protocols
Motor Imagery BCI Protocol:
- Task Design: Implement left vs. right hand motor imagery trials (25 each, randomized)
- Visual Cueing: Present directional arrows (5s duration) following rest period (20s) [15]
- Sensor Placement: Focus on C3, C4 positions over motor cortex with additional prefrontal coverage [15]
- Feature Extraction:
- Classification: Utilize support vector machines (SVM) with early temporal features for rapid BCI response [15]
Mental Workload/Cognitive BCI Protocol:
- Task Design: Implement n-back tasks (0-back vs. 2-back) or mental arithmetic vs. baseline [16]
- Sensor Placement: Emphasize prefrontal cortex coverage [5]
- Feature Extraction:
- Classification: Apply linear discriminant analysis (LDA) or deep learning approaches [5] [17]
Advanced Integration Methodologies
Spatial-Temporal Alignment Network (STA-Net): Advanced deep learning approaches address the inherent temporal misalignment between EEG and fNIRS signals. The STA-Net architecture includes:
- fNIRS-guided Spatial Alignment (FGSA): Uses fNIRS spatial information to identify sensitive brain regions and weight corresponding EEG channels [14]
- EEG-guided Temporal Alignment (EGTA): Generates temporally aligned fNIRS signals using cross-attention mechanisms to compensate for hemodynamic delay [14]
- This approach has demonstrated classification accuracies of 69.65% for motor imagery, 85.14% for mental arithmetic, and 79.03% for word generation tasks [14]
Decision-Level Fusion with Uncertainty Modeling:
- Implement Dirichlet distribution parameter estimation to model uncertainty in unimodal decisions [17]
- Apply Dempster-Shafer Theory for two-layer reasoning to fuse evidence from both modalities [17]
- This approach has achieved 83.26% accuracy in motor imagery classification, outperforming single-modality implementations [17]
The integration of EEG and fNIRS technologies presents a powerful approach to overcoming the fundamental trade-off between temporal and spatial resolution in non-invasive brain imaging. Through strategic multimodal fusion, researchers can leverage EEG's millisecond-level temporal resolution alongside fNIRS's superior spatial localization capabilities, enabling more robust and accurate BCIs. The experimental protocols and methodological considerations outlined in this application note provide a framework for optimizing simultaneous EEG-fNIRS setups, with particular relevance for motor imagery and cognitive task classification in BCI research. As integration methodologies continue to advance, particularly through deep learning approaches that explicitly address spatial-temporal alignment challenges, hybrid EEG-fNIRS systems are poised to significantly enhance the performance and practical applicability of next-generation brain-computer interfaces.
The integration of Electroencephalography (EEG) and functional Near-Infrared Spectroscopy (fNIRS) represents a transformative approach in brain-computer interface (BCI) research, offering a more comprehensive view of brain function than either modality could provide alone [18]. This synergistic framework capitalizes on the complementary strengths of both techniques: EEG provides excellent temporal resolution on the millisecond scale, capturing rapid neural electrical activity, while fNIRS offers valuable spatial information regarding brain activation by localizing hemodynamic responses associated with neuronal activity [18] [2]. Simultaneous EEG-fNIRS recordings bridge a critical gap in neuroimaging, enabling researchers to correlate the fast dynamics of electrophysiological activity with the slower hemodynamic changes that reflect metabolic demands of neural processing [19].
The fundamental synergy stems from measuring different aspects of brain function: EEG records electrical potentials generated by synchronized neuronal firing, while fNIRS measures concentration changes in oxygenated (HbO) and deoxygenated hemoglobin (HbR) in the blood, which serve as proxies for neural metabolic activity [20]. This complementary relationship makes the combined approach particularly valuable for investigating complex cognitive processes, developing more robust BCIs, and advancing clinical applications in neurology and psychiatry [2] [21].
Fundamental Principles and Complementary Nature
Technical Basis of EEG and fNIRS
Electroencephalography (EEG) detects electrical activity generated by the synchronized firing of neuronal populations beneath the scalp. With a temporal resolution in the millisecond range, EEG excels at tracking the rapid dynamics of brain activity but suffers from limited spatial resolution due to signal attenuation and smearing as electrical potentials pass through various tissues before reaching scalp electrodes [2] [19].
Functional Near-Infrared Spectroscopy (fNIRS) employs near-infrared light to measure cortical brain activity by detecting hemodynamic responses associated with neuronal activity. Light in the 700-900 nm range is shone through the scalp, and detectors measure backscattered light, allowing calculation of concentration changes in oxygenated and deoxygenated hemoglobin based on differential absorption characteristics [20]. While fNIRS provides superior spatial resolution compared to EEG (approximately 2 cm depth), its temporal resolution is limited by the inherent speed of the hemodynamic response, which typically unfolds over seconds [19].
The Synergistic Relationship
The complementary characteristics of EEG and fNIRS create a powerful synergistic relationship for studying brain function, as illustrated in the following diagram:
This synergy enables researchers to investigate the relationship between electrical brain activity and subsequent metabolic responses, providing a more complete picture of neurovascular coupling—the fundamental process linking neural activity to cerebral blood flow changes [2]. The combined approach is particularly advantageous for BCI applications, where understanding both the timing and spatial distribution of brain activity is essential for accurate classification of user intent [18] [17].
Applications in Brain-Computer Interface Research
Motor Imagery and Execution
Motor imagery (MI) represents one of the most widely investigated applications for hybrid EEG-fNIRS BCI systems. Research has demonstrated that combining temporal EEG patterns with spatial fNIRS activation profiles significantly improves classification accuracy of imagined movements. A recent study utilizing deep learning and evidence theory for EEG-fNIRS signal integration achieved an average accuracy of 83.26% for MI classification, representing a 3.78% improvement over state-of-the-art unimodal methods [17]. This enhanced performance stems from the complementary information provided by each modality: EEG captures event-related desynchronization/synchronization during motor imagery, while fNIRS detects hemodynamic changes in the motor cortex [18].
Cognitive Monitoring and Semantic Decoding
Hybrid EEG-fNIRS systems show considerable promise for decoding semantic information during cognitive tasks. Recent research has successfully differentiated between semantic categories (animals vs. tools) during silent naming and sensory-based imagery tasks using simultaneous EEG-fNIRS recordings [19]. The experimental paradigm included:
- Silent naming: Participants silently named displayed objects
- Visual imagery: Mental visualization of objects
- Auditory imagery: Imagination of sounds associated with objects
- Tactile imagery: Imagination of touching objects
This approach to semantic neural decoding could enable more intuitive BCIs that communicate conceptual meaning directly, bypassing the character-by-character spelling used in current systems [19].
Clinical and Diagnostic Applications
The integration of EEG and fNIRS has demonstrated significant potential across various clinical domains, leveraging their complementary strengths for improved diagnosis and monitoring:
Table: Clinical Applications of EEG-fNIRS Integration
| Clinical Domain | Application | Benefits of EEG-fNIRS Integration |
|---|---|---|
| Disorders of Consciousness | Detecting neural signatures of cognitive processes | Combines EEG's sensitivity to transient changes with fNIRS's spatial localization of active regions [21] [22] |
| Stroke Rehabilitation | Motor function recovery training | Provides comprehensive feedback on both electrical and hemodynamic responses during therapy [22] [23] |
| ADHD | Training inhibitory control and working memory | Enables monitoring of both rapid neural oscillations and sustained prefrontal activation [2] [22] |
| Epilepsy | Seizure focus localization and monitoring | Correlates ictal electrical discharges with localized hemodynamic changes [2] |
| Anesthesia Monitoring | Depth of anesthesia assessment | Combines EEG-based anesthetic depth indicators with fNIRS cerebral oxygenation monitoring [2] |
Experimental Protocols and Methodologies
Protocol 1: Semantic Category Decoding
This protocol outlines the methodology for differentiating between semantic categories (animals vs. tools) using simultaneous EEG-fNIRS recordings during various mental imagery tasks [19].
Participant Preparation and Equipment
- Participants: Recruit right-handed native speakers (for language-dependent tasks) with normal or corrected-to-normal vision. Sample size: 12 participants for combined EEG-fNIRS recording.
- EEG System: 64-channel system with sampling rate ≥1000 Hz, electrode impedances maintained below 5 kΩ.
- fNIRS System: Continuous-wave system with laser sources, avalanche photodiode detectors, sampling rate ≥62.5 Hz, measuring HbO and HbR concentration changes.
- Synchronization: Use photoelectric marking or hardware synchronization to align EEG and fNIRS data streams with sub-second precision.
Stimuli and Task Design
- Stimulus Set: 18 animals and 18 tools selected for recognizability and suitability across mental tasks. Images converted to grayscale, cropped to 400×400 pixels, and presented against white background.
- Task Conditions:
- Silent Naming: Participants silently name the displayed object in their mind.
- Visual Imagery: Participants visualize the object in their mind.
- Auditory Imagery: Participants imagine sounds associated with the object.
- Tactile Imagery: Participants imagine the feeling of touching the object.
- Trial Structure: Each trial consists of (1) stimulus presentation (2-3 seconds), (2) mental task period (3-5 seconds), and (3) rest interval (5+ seconds). Task order randomized across blocks.
Data Acquisition Procedure
- Apply EEG cap and fNIRS probes according to manufacturer specifications, ensuring proper contact and signal quality.
- Position fNIRS sources and detectors to cover frontal, temporal, and parietal regions based on the hypothesized neural correlates of semantic processing.
- Conduct a brief practice session to familiarize participants with tasks.
- Record simultaneous baseline activity during rest condition.
- Present stimuli in randomized order across task conditions, with adequate rest periods between blocks to minimize fatigue.
- Monitor data quality throughout acquisition, checking for artifacts and signal integrity.
The following diagram illustrates the experimental workflow:
Protocol 2: Conflict Processing Using Stroop Task
This protocol details the simultaneous EEG-fNIRS recording during Stroop task performance, a classic paradigm for investigating conflict monitoring and processing [24].
Experimental Setup
- Participants: 21 healthy volunteers with normal color vision, no history of neurological or psychiatric disorders.
- EEG Configuration: 34-channel recording according to 10-20 system, sampling rate 1000 Hz, band-pass filter 0.05-100 Hz, 50 Hz notch filter.
- fNIRS Configuration: 20-channel system (4 sources, 16 detectors) covering frontal and parietal lobes, sampling rate 100 Hz, measuring Δ[HbO] and Δ[Hb] at 785 nm and 850 nm wavelengths.
- Stimuli: Chinese color-word matching task with neutral and incongruent conditions.
Task Procedure
- Neutral Stimuli: Upper character is a non-color word presented in colored ink; lower character is a color word presented in white.
- Incongruent Stimuli: Upper character is a color word presented in a different colored ink.
- Task: Participants judge whether the color of the upper character matches the meaning of the lower character.
- Response: Left mouse button for match, right button for non-match.
- Block Design: 4 blocks (neutral, neutral, incongruent, incongruent), each with 16 trials.
- Trial Structure: Stimulus presentation (2 seconds) followed by inter-trial interval (5 seconds).
- Timing: 30-second rest periods before first block and after last block.
Data Quality Assurance
- Maintain EEG electrode impedances below 5 kΩ throughout recording.
- Verify fNIRS signal quality by checking raw intensity levels before formal experiment.
- Use photoelectric marking system to synchronize stimulus presentation with data acquisition.
- Monitor for movement artifacts and provide reminders to minimize motion.
Data Processing and Fusion Approaches
The integration of EEG and fNIRS data can be accomplished at multiple levels, each with distinct advantages:
Table: Data Fusion Approaches for EEG-fNIRS Integration
| Fusion Level | Description | Methods | Applications |
|---|---|---|---|
| Hardware Level | Physical integration of EEG electrodes and fNIRS optodes | Customized helmets, 3D-printed mounts, elastic caps with integrated components [2] | All simultaneous recording paradigms |
| Data Level | Direct combination of raw or preprocessed signals | Common average referencing, joint filtering, tensor-based fusion [18] | Signal quality enhancement, artifact removal |
| Feature Level | Concatenation or transformation of extracted features | STFT for EEG time-frequency images + fNIRS spectral entropy [18] | Motor imagery classification, cognitive state monitoring |
| Decision Level | Fusion of classification outcomes from separate models | Dempster-Shafer theory, weighted voting, Bayesian fusion [17] | Semantic decoding, conflict processing, clinical diagnosis |
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful implementation of simultaneous EEG-fNIRS experiments requires careful selection of equipment and materials. The following table details key components and their functions:
Table: Essential Materials for Simultaneous EEG-fNIRS Research
| Category | Item | Specifications | Function/Purpose |
|---|---|---|---|
| EEG System | Amplifier and Electrodes | 34+ channels, sampling rate ≥1000 Hz, impedance <5 kΩ [24] | Records electrical brain activity with high temporal resolution |
| fNIRS System | Sources and Detectors | 2 wavelengths (785nm, 850nm), sampling rate ≥62.5 Hz [20] | Measures hemodynamic responses via light absorption |
| Integration Hardware | Custom Helmet | 3D-printed or thermoplastic custom fit [2] | Maintains precise, stable positioning of both modalities |
| Synchronization | Photoelectric Marker | Light-sensitive trigger device | Ensures temporal alignment of EEG and fNIRS data |
| Software | Analysis Package | MATLAB, Python, NIRS-SPM, Homer2 | Preprocessing, feature extraction, data fusion |
| Stimulus Presentation | Display Software | PsychToolbox, Presentation, E-Prime | Controls experimental paradigm timing |
| Quality Assurance | Impedance Checker | Electrode tester <5 kΩ | Verifies EEG signal quality at recording start |
Advanced Data Analysis and Fusion Methodologies
Multimodal DenseNet Fusion (MDNF) Model
Recent advances in deep learning have yielded sophisticated frameworks for integrating EEG and fNIRS data. The Multimodal DenseNet Fusion (MDNF) model represents a cutting-edge approach that transforms EEG data into two-dimensional time-frequency images using Short-Time Fourier Transform (STFT), then applies transfer learning to extract discriminative features which are integrated with fNIRS-derived spectral entropy features [18]. This architecture effectively bridges the gap between temporal richness of EEG and spatial specificity of fNIRS, demonstrating superior classification accuracy across various cognitive and motor imagery tasks [18].
The MDNF implementation involves:
- EEG Transformation: Convert raw EEG signals to 2D spectrogram images via STFT
- Feature Extraction: Utilize pre-trained DenseNet for automated feature learning from EEG spectrograms
- fNIRS Processing: Compute spectral entropy features from hemodynamic signals
- Feature Fusion: Integrate EEG and fNIRS features in a unified representation space
- Classification: Employ fully connected layers for task-specific prediction
Evidence Theory for Decision Fusion
An innovative end-to-end signal fusion method combines deep learning with Dempster-Shafer Theory (DST) for motor imagery classification [17]. This approach includes:
- EEG Pathway: Spatiotemporal feature extraction using dual-scale temporal convolution and depthwise separable convolution, enhanced with hybrid attention mechanisms
- fNIRS Pathway: Spatial convolution across channels to explore regional activation differences, combined with parallel temporal convolution and gated recurrent units (GRU)
- Decision Fusion: Uncertainty quantification via Dirichlet distribution parameter estimation, followed by two-layer reasoning using DST to fuse evidence from both modalities
This methodology has demonstrated state-of-the-art performance, achieving 83.26% accuracy in MI classification tasks [17].
Implementation Considerations and Technical Challenges
Hardware Integration and Signal Quality
Successful simultaneous EEG-fNIRS recording requires addressing several technical challenges:
- Probe Placement: Strategic selection of EEG channels based on neuroanatomical locations and their associations with cognitive and motor functions is essential [18]. Similarly, fNIRS optode placement must cover regions of interest while minimizing interference with EEG electrodes.
- Cross-Modal Artifacts: Physiological artifacts (e.g., cardiac pulsation, respiration) affect both modalities differently and require specialized filtering approaches.
- Synchronization: Precise temporal alignment of EEG and fNIRS data streams is critical. Hardware synchronization provides the most accurate alignment, while software methods offer more flexibility [2].
- Comfort and Stability: Extended recordings require comfortable, stable probe placement to minimize motion artifacts. Customized helmets using 3D printing or thermoplastic materials can improve fit and stability [2].
Data Processing Pipelines
Effective data processing requires modality-specific approaches:
- EEG Processing: Filtering (0.5-50 Hz bandpass, 50/60 Hz notch), artifact removal (ocular, cardiac, motion), segmentation, and feature extraction (time-domain, frequency-domain, time-frequency representations)
- fNIRS Processing: Conversion of raw light intensity to optical density, motion artifact correction, bandpass filtering (0.01-0.5 Hz) to remove physiological noise, conversion to hemoglobin concentration changes via modified Beer-Lambert law
- Multimodal Integration: Temporal alignment, common referencing, and application of fusion algorithms at appropriate levels (data, feature, or decision fusion)
The following diagram illustrates a comprehensive data processing workflow:
Historical Development and Commercial Evolution of Hybrid EEG-fNIRS Systems
The integration of electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS) represents a paradigm shift in non-invasive neuroimaging, creating a powerful hybrid modality for brain-computer interface (BCI) research. This fusion addresses fundamental limitations inherent in each standalone technique: EEG provides excellent temporal resolution but suffers from poor spatial localization, while fNIRS offers superior spatial resolution but slower temporal response [2]. The complementary nature of these modalities has catalyzed their integration, enabling unprecedented insights into brain function across diverse clinical and research applications.
The evolution of hybrid EEG-fNIRS systems has progressed from preliminary proof-of-concept studies to sophisticated, commercially viable platforms. This journey has been marked by significant interdisciplinary collaboration between engineers, neuroscientists, and clinicians. Modern systems now demonstrate robust synchronization capabilities, user-friendly interfaces, and expanding applications in both laboratory and real-world settings [25]. The commercial landscape has similarly evolved, with increasing numbers of manufacturers offering integrated solutions that cater to the growing demand for multimodal brain imaging in research and therapeutic applications.
Historical Development and Technological Evolution
Early Foundations and Conceptual Framework
The historical development of hybrid EEG-fNIRS systems emerged from the recognized need to overcome limitations of unimodal brain imaging approaches. Initial research in the early 2000s focused on establishing the technical feasibility of simultaneous acquisition, addressing fundamental challenges related to hardware integration and signal synchronization [25]. These pioneering studies demonstrated that electrophysiological (EEG) and hemodynamic (fNIRS) measurements could be successfully co-registered, providing complementary information about brain activity.
The conceptual framework for hybridization was formally articulated by Pfurtscheller et al. (2010), who established four essential criteria for genuine hybrid systems: (1) direct acquisition of brain activity, (2) utilization of multiple brain signal acquisition modalities, (3) real-time signal processing for communication between brain and computer, and (4) provision of feedback outcomes [25]. This framework distinguished true hybrid BCIs from simple multi-modal recording setups and established standards for the field's development.
Hardware Integration Milestones
The evolution of hardware integration has progressed through several distinct phases, each addressing critical technical challenges:
Initial Solutions (Pre-2010): Early systems utilized separate EEG and fNIRS equipment with post-hoc synchronization, resulting in temporal alignment limitations. Researchers often adapted existing EEG caps by manually creating openings for fNIRS optodes, leading to suboptimal contact pressure and positioning consistency [2].
Dedicated Hybrid Caps (2010-2017): The development of purpose-built integration caps represented a significant advancement. Initial commercial offerings used elastic fabrics with predefined openings for both electrode and optode placement, improving reproducibility but still facing challenges with maintaining consistent optode-scalp contact pressure across different head shapes [2].
Advanced Customization (2017-Present): Recent innovations incorporate 3D printing and thermoplastic materials to create subject-specific helmets that optimize probe placement and contact pressure. These customized solutions enhance signal quality but at increased cost and complexity [2]. Modern commercial systems now offer integrated caps with optimized layouts for specific applications, such as motor imagery or prefrontal cortex monitoring.
Synchronization Techniques Evolution
The progression of synchronization methods has been crucial for effective data fusion:
Software-based Synchronization: Early approaches used software triggers and shared clock systems between separate devices, achieving synchronization at the tens of milliseconds level, sufficient for fNIRS but suboptimal for high-temporal-resolution EEG analysis [2].
Hardware-level Integration: Advanced systems implemented unified processors that simultaneously handle EEG and fNIRS input/output, achieving microsecond-level synchronization precision. This integration enables truly simultaneous data acquisition essential for investigating neurovascular coupling dynamics [2].
Modern Commercial Systems: Contemporary commercial platforms incorporate dedicated synchronization modules with hardware triggers and shared analog-digital converters, ensuring temporal alignment sufficient for analyzing complex inter-modal relationships during cognitive tasks [26].
Table 1: Evolution of Hybrid EEG-fNIRS System Capabilities
| Time Period | Primary Integration Method | Synchronization Precision | Key Commercial Developments |
|---|---|---|---|
| 2005-2010 | Modified EEG caps with fNIRS openings | 50-100 ms (software triggers) | First research prototypes; Custom solutions |
| 2011-2017 | Dedicated hybrid caps (elastic fabric) | 10-50 ms (improved triggers) | Early commercial systems (NIRx, Artinis with EEG partners) |
| 2018-2023 | 3D-printed custom interfaces | 1-5 ms (hardware synchronization) | Integrated commercial platforms (g.tec hybrid systems) |
| 2024-Present | Subject-specific optimized layouts | <1 ms (unified processors) | Commercial systems with real-time analysis capabilities |
Technical Specifications and Performance Metrics
Comparative Modal Characteristics
The fundamental rationale for EEG-fNIRS integration stems from their complementary characteristics across multiple dimensions of measurement. EEG records electrical potentials generated by synchronized neuronal activity with millisecond temporal resolution, ideal for capturing rapid neural dynamics during cognitive tasks or in response to stimuli [2]. However, the electrical signals are attenuated and distorted by passage through cerebrospinal fluid, skull, and scalp, resulting in limited spatial resolution of approximately 2-3 cm under optimal conditions [19].
Conversely, fNIRS measures hemodynamic responses associated with neural activity through light absorption changes in oxygenated and deoxygenated hemoglobin. While slower in temporal response (typically 2-10 Hz sampling versus 256-1000 Hz for EEG), fNIRS provides superior spatial localization (5-10 mm resolution) and direct measurement of regional brain activation [26]. fNIRS is less susceptible to movement artifacts and electromagnetic interference, making it suitable for more naturalistic environments [2].
The neurovascular coupling relationship—the biological link between neural activity and subsequent hemodynamic response—forms the physiological basis for correlating EEG and fNIRS signals. This relationship exhibits complex temporal dynamics that vary across brain regions and cognitive states, with hemodynamic responses typically lagging 4-8 seconds behind electrical activity [27].
Table 2: Performance Comparison of Neuroimaging Modalities
| Parameter | EEG | fNIRS | Hybrid EEG-fNIRS | fMRI |
|---|---|---|---|---|
| Temporal Resolution | Millisecond level | ~2-10 Hz | Millisecond (EEG) + ~2-10 Hz (fNIRS) | 0.5-2 Hz |
| Spatial Resolution | 2-3 cm | 5-10 mm | 5-10 mm (fNIRS-guided) | 1-3 mm |
| Portability | High | High | Moderate-high | Low |
| Cost | Low-moderate | Moderate | Moderate | High |
| Artifact Resistance | Low (electrical interference) | Moderate (movement) | Moderate (complementary) | High |
| Measurement Depth | Cortical surface | Superficial cortex (2-3 cm) | Superficial cortex | Whole brain |
Commercial System Evolution
The commercial landscape for hybrid EEG-fNIRS systems has expanded significantly, with several manufacturers now offering integrated solutions:
Early Commercialization (2010-2017): Initial commercial offerings focused on compatibility between existing EEG and fNIRS systems from the same manufacturer or partners. These systems required significant technical expertise to operate and synchronize effectively [25].
Current Integrated Systems (2018-Present): Modern commercial systems feature unified hardware platforms with simplified user interfaces. Examples include g.tec's hybrid systems with integrated amplifiers and NirScan's combined acquisition units [26]. These systems typically incorporate 32-64 EEG channels alongside 16-64 fNIRS channels, with sampling rates up to 1000 Hz for EEG and 10-50 Hz for fNIRS.
Performance Metrics: Contemporary systems achieve classification accuracies of 80-95% for various BCI tasks, representing 5-15% improvement over unimodal approaches [18] [7]. The development of standardized communication protocols (Lab Streaming Layer) has facilitated integration of equipment from different manufacturers, increasing flexibility for researchers.
Experimental Protocols and Methodologies
Protocol 1: Motor Imagery for BCI Applications
Motor imagery (MI) protocols represent one of the most established applications for hybrid EEG-fNIRS systems, particularly in rehabilitation and assistive technology development.
Participant Preparation and Setup
Participant Selection: Recruit right-handed participants (to minimize hemispheric dominance variability) with normal or corrected-to-normal vision. For clinical studies, include both healthy controls and patient populations (e.g., intracerebral hemorrhage patients) with appropriate consent procedures [26].
Equipment Setup: Use a customized hybrid cap with 32 EEG electrodes positioned according to the international 10-20 system and 32 fNIRS sources with 30 detectors creating 90 measurement channels through source-detector pairing at 3 cm separation distances [26]. Ensure proper scalp coupling through hair parting and application of appropriate conductive (EEG) and optical (fNIRS) gels.
Signal Quality Verification: Check EEG impedance levels (<10 kΩ) and fNIRS scalp coupling index (SCI > 0.7) before beginning experimental tasks. Reject channels failing quality thresholds from subsequent analysis [26].
Experimental Paradigm
Baseline Recording: Acquire 1-minute eyes-closed followed by 1-minute eyes-open baseline measurements, demarcated by auditory cues (200 ms beep) [26].
Task Structure: Implement a trial-based design with the following sequence:
- Visual Cue (2 s): Display a directional arrow (left/right) indicating the required motor imagery.
- Execution Phase (10 s): Present a central fixation cross following an auditory cue (200 ms beep). Participants perform kinesthetic motor imagery of grasping with the indicated hand at approximately 1 imagined grasp per second.
- Inter-trial Rest (15 s): Blank screen for rest and return to baseline [26].
Session Structure: Conduct multiple sessions with at least 30 trials each (15 left/right hand MI), with intersession breaks adjusted based on participant fatigue.
Data Acquisition Parameters
- EEG Settings: Sampling rate 256-1000 Hz, bandpass filter 0.5-45 Hz, recording reference at Cz or average reference.
- fNIRS Settings: Sampling rate 10-12.5 Hz, wavelengths 760 nm and 850 nm to measure oxygenated (HbO) and deoxygenated hemoglobin (HbR) concentration changes.
- Synchronization: Use event markers from stimulus presentation software (e.g., E-Prime) transmitted simultaneously to both recording systems [26].
Protocol 2: Semantic Neural Decoding
Semantic decoding protocols investigate the neural representation of conceptual knowledge, with applications to advanced communication BCIs.
Stimuli and Task Design
Stimulus Selection: Utilize images representing distinct semantic categories (e.g., 18 animals and 18 tools) selected for suitability across multiple mental tasks. Convert images to grayscale, crop to 400×400 pixels, and present against white background [19].
Mental Tasks: Implement four distinct mental tasks in randomized order across blocks:
- Silent Naming: Participants silently name the displayed object in their native language.
- Visual Imagery: Participants visualize the object in their minds.
- Auditory Imagery: Participants imagine sounds associated with the object.
- Tactile Imagery: Participants imagine the feeling of touching the object [19].
Trial Structure: Each trial consists of stimulus presentation (3-5 s) followed by the mental task period (3-5 s), with inter-trial intervals of 10-15 s.
Data Collection Parameters
- EEG Configuration: 30 electrodes positioned according to the international 10-5 system, sampling rate 1000 Hz (downsampled to 200 Hz) [19].
- fNIRS Configuration: 36 channels (14 sources, 16 detectors) with 30 mm inter-optode distance, following the 10-20 system, sampling rate 12.5 Hz (downsampled to 10 Hz), wavelengths 760 nm and 850 nm [19].
- Participant Instructions: Emphasize minimizing physical movements (eye movements, facial expressions, head/body motions) during mental tasks to reduce artifacts.
Data Processing and Analysis Framework
A standardized processing pipeline ensures reproducible analysis of hybrid EEG-fNIRS data:
Preprocessing Steps
EEG Preprocessing: Apply bandpass filtering (0.5-45 Hz), remove line noise, re-reference to average reference, detect and reject artifacts using independent component analysis (ICA), particularly for ocular and muscle artifacts.
fNIRS Preprocessing: Convert raw intensity to optical density, apply bandpass filtering (0.01-0.2 Hz for task-related signals), detect motion artifacts using moving standard deviation or peak-to-peak amplitude thresholds, correct using wavelet-based or PCA-based methods [28]. For systemic physiological noise removal, employ short-separation regression (if available) or principal component filtering [28].
Temporal Alignment: Precisely align EEG and fNIRS data using synchronization triggers, accounting for inherent hemodynamic delay (typically 4-8 seconds) in fNIRS responses relative to EEG.
Feature Extraction and Fusion
EEG Features: Extract time-frequency features using wavelet transform or short-time Fourier transform, particularly in motor imagery paradigms focusing on sensorimotor rhythms (8-30 Hz). For cognitive tasks, extract event-related potentials (ERPs) or power in specific frequency bands.
fNIRS Features: Calculate oxygenated (HbO) and deoxygenated (HbR) hemoglobin concentration changes using modified Beer-Lambert law. Extract features including mean, peak, slope, and area under the curve during task periods.
Feature Fusion: Implement either early fusion (concatenating features before classification) or late fusion (combining classifier decisions) approaches. For motor imagery, combining EEG band power with fNIRS HbO concentrations typically yields optimal results [18].
Classification Approaches
Traditional Machine Learning: Utilize support vector machines (SVM), linear discriminant analysis (LDA), or random forests with carefully selected feature combinations.
Deep Learning Architectures: Implement multimodal denseNet fusion (MDNF) models that transform EEG data into 2D time-frequency representations using short-time Fourier transform and combine with fNIRS spectral entropy features [18].
Ensemble Methods: Apply stacking ensemble learning combining multiple classifiers (Naïve Bayes, SVM, Random Forest, k-NN) with genetic algorithm-based feature selection [7].
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Essential Components for Hybrid EEG-fNIRS Research
| Component Category | Specific Items | Function/Purpose | Technical Specifications |
|---|---|---|---|
| Acquisition Hardware | Hybrid EEG-fNIRS cap | Provides structural foundation for electrode/optode placement | 32-64 EEG channels, 16-64 fNIRS channels, international 10-20 system compliance |
| EEG amplifier | Records electrical brain activity | 24-64 channels, sampling rate ≥256 Hz, input impedance >100 MΩ | |
| fNIRS system | Measures hemodynamic responses | 2+ wavelengths (760, 850 nm), sampling rate ≥10 Hz, source-detector separation 30 mm | |
| Synchronization module | Ensures temporal alignment of multimodal data | Hardware triggers, shared clock, <1 ms precision | |
| Disposable Supplies | EEG electrolyte gel | Ensures electrical conductivity between scalp and electrodes | Chloride-based, low impedance, non-irritating |
| fNIRS optical gels | Improves light coupling between optodes and scalp | High refractive index matching, non-toxic | |
| Abrasive prep gel | Gentle scalp exfoliation to reduce impedance | Mild abrasive particles in electrolyte solution | |
| Disposable electrode disks | Single-use EEG electrodes | Ag/AgCl composition, adhesive backing | |
| Stimulus Presentation | Presentation software | Controls experimental paradigm timing | Precision timing, trigger output, E-Prime, PsychoPy, or Presentation |
| Display monitor | Visual stimulus presentation | High refresh rate (≥120 Hz), minimal latency | |
| Response collection device | Records participant responses | Button boxes, keyboards, or specialized input devices | |
| Data Analysis Tools | Signal processing software | Preprocessing and analysis of multimodal data | MATLAB with EEGLAB, NIRS-KIT, Homer2, MNE-Python |
| Classification libraries | Machine learning implementation | scikit-learn, TensorFlow, PyTorch with custom hybrid BCI extensions | |
| Calibration & Quality Assurance | Impedance checker | Verifies EEG electrode-scalp contact | <10 kΩ threshold for acceptable connections |
| Optical power meter | Validates fNIRS source output | Measures intensity at optode tips | |
| Phantom test objects | System validation and calibration | Tissue-simulating materials with known optical properties |
Commercial Applications and Future Directions
The commercial evolution of hybrid EEG-fNIRS systems has expanded beyond research laboratories into various practical applications. Current commercial systems demonstrate robust performance in clinical neurodiagnostics, neurorehabilitation, and consumer neuroscience applications.
Emerging Commercial Applications
Clinical Rehabilitation: Hybrid systems show particular promise in stroke rehabilitation, with systems specifically validated for intracerebral hemorrhage patients demonstrating the ability to track motor recovery through combined electrophysiological and hemodynamic monitoring [26]. Commercial systems are increasingly incorporating patient-specific adaptation algorithms to accommodate pathological neurovascular coupling.
Assistive Communication: The development of semantic decoding BCIs using hybrid systems offers potential for more natural communication interfaces, moving beyond character-by-character spelling to direct concept communication [19]. Commercial entities are exploring these approaches for locked-in syndrome and other severe communication impairments.
Neuromarketing and Consumer Research: The commercial sector has adopted hybrid systems for evaluating consumer responses to products and advertisements, leveraging the comprehensive brain activity assessment provided by combined electrical and hemodynamic monitoring.
Future Commercial Directions
The future commercial evolution of hybrid EEG-fNIRS systems will likely focus on several key areas:
Miniaturization and Wearability: Next-generation systems are transitioning toward more compact, wearable designs suitable for real-world monitoring outside laboratory environments. This includes developments in wireless technology, battery life optimization, and ergonomic design.
Real-Time Processing Integration: Commercial systems are increasingly incorporating real-time analysis capabilities, enabling immediate feedback for neurorehabilitation and BCI applications. Advanced processors and optimized algorithms allow complex multimodal fusion with minimal latency.
Standardization and Interoperability: Industry-wide standards for data formats, synchronization protocols, and interface specifications are emerging to facilitate multi-site studies and technology transfer between research and clinical applications.
AI-Enhanced Analytics: Commercial systems are beginning to integrate artificial intelligence and machine learning directly into acquisition platforms, providing automated interpretation and reducing the expertise required for system operation and data analysis.
The continued commercial evolution of hybrid EEG-fNIRS systems promises to further bridge the gap between laboratory research and practical applications, ultimately expanding access to sophisticated brain monitoring technologies across diverse fields including clinical medicine, neuroscience research, and human-computer interaction.
Building and Implementing a Hybrid EEG-fNIRS BCI: From Hardware Setup to Data Fusion
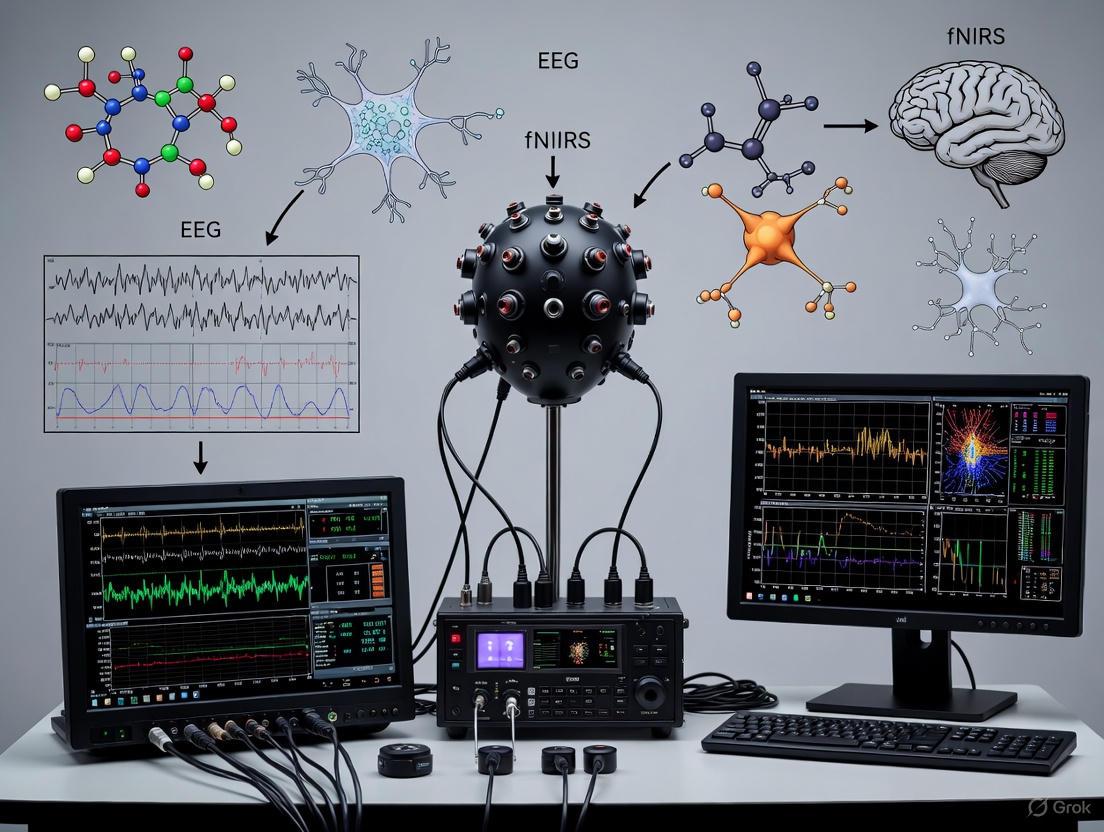
This document provides detailed application notes and protocols for the design and implementation of a unified helmet-based acquisition system for simultaneous electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS). The integration of these two non-invasive neuroimaging modalities leverages their complementary strengths to advance brain-computer interface (BCI) research. EEG provides millisecond-level temporal resolution of electrical brain activity, while fNIRS offers superior spatial localization for hemodynamic responses, enabling the development of more robust and accurate hybrid BCI systems [29] [30]. The helmet form factor is critical for ensuring consistent sensor placement, user comfort, and mobility, which are essential for conducting valid and reproducible experiments in both clinical and laboratory settings [26].
System Architecture and Integration Principles
The core of a unified EEG-fNIRS system is a co-located, modular sensor platform integrated into a single helmet. The design must facilitate simultaneous data acquisition from both modalities with precise temporal synchronization.
Key Integration Principles:
- Co-location of Sensors: EEG electrodes and fNIRS optodes must be positioned on the scalp to cover the same cortical regions of interest, particularly the motor cortex for motor imagery (MI) paradigms [26] [30]. This allows for the direct correlation of electrical and hemodynamic activity from the same brain area.
- Temporal Synchronization: A common hardware trigger must initialize both the EEG amplifier and the fNIRS system to ensure data streams are aligned with millisecond precision. This is often achieved via a synchronization pulse from the stimulus presentation software (e.g., E-Prime) sent to both acquisition devices [26].
- * form factor and Ergonomics:* The helmet should be designed for a secure and comfortable fit to minimize motion artifacts. Considerations include total weight, center of gravity, and the use of customizable, padded liners to accommodate different head sizes and shapes [31].
Table 1: Core System Components and Specifications
| Component | Key Specifications | Integration Role |
|---|---|---|
| EEG Amplifier [26] | ≥ 32 channels; Sampling Rate: ≥ 256 Hz; Input Referenced | Captures electrical potentials from the scalp with high temporal resolution. |
| fNIRS System [26] | Continuous-wave; Sampling Rate: ~11 Hz; Lasers & Photodetectors | Measures hemodynamic changes (HbO/HbR) via near-infrared light. |
| Hybrid EEG-fNIRS Cap [26] | Integrated 32-electrode & 62-optode (32 sources, 30 detectors) layout. | Provides fixed, co-located geometry for sensors; ensures coverage of target cortices. |
| Stimulus Presentation Software [26] | e.g., E-Prime; Capable of sending event markers. | Presents experimental paradigm and outputs synchronization pulses for data alignment. |
The logical data acquisition and synchronization workflow is outlined below.
The Researcher's Toolkit: Essential Materials and Reagents
A successful experimental setup requires specific materials for sensor integration, signal quality assurance, and participant safety.
Table 2: Essential Research Reagents and Materials
| Item | Function / Purpose | Application Notes |
|---|---|---|
| Conductive EEG Gel (e.g., NeuroPrep Gel) [30] | Reduces impedance between the scalp and EEG electrodes by filling irregularities. | Applied via blunt-tipped syringe. Essential for high-fidelity signal acquisition but requires post-session cleaning. |
| Ten20 Paste [30] | An alternative conductive medium for securing EEG electrodes and maintaining low impedance. | Offers stable connectivity for longer recording sessions. |
| Abrasive Skin Prep Gel | Gently exfoliates the scalp skin at electrode sites to remove dead skin cells and oils. | Significantly lowers initial skin impedance, improving signal quality. |
| Isopropyl Alcohol (70%) | Cleanses the scalp and hair before EEG setup and removes conductive gel post-session. | Ensures a clean interface for sensors and maintains hygiene. |
| Blunt-Tipped Syringes | Precise application of conductive gel onto individual EEG electrodes within the cap. | Preents gel bridging between adjacent electrodes, which can cause signal shorts. |
| fNIRS Optode Holders | Securely positions optical sources and detectors on the scalp at a fixed distance (typically 3 cm). | Integrated into the hybrid cap design to maintain optimal source-detector separation for penetration depth. |
| Disposable ECG Electrodes | Can be used as ground or reference electrodes for the EEG system. | Placed on bony landmarks (e.g., mastoid). |
Experimental Protocol: A Standard Motor Imagery Paradigm
The following protocol details a classic left-hand/right-hand motor imagery task, a common paradigm in BCI research for developing motor restoration applications [26] [30].
4.1. Participant Preparation and Setup
- Informed Consent: Obtain written informed consent approved by an institutional ethics committee (e.g., TJ-IRB202412123 [26]).
- Helmet Fitting: Measure the participant's head circumference and select the appropriate hybrid cap size (e.g., Model M for 54-58 cm [26]). Securely fit the cap, ensuring the Cz electrode is aligned with the vertex of the head.
- EEG Preparation: For each electrode, part the hair and abrade the skin with preparatory gel. Fill the electrodes with conductive gel using a blunt-tipped syringe until impedances are stabilized below 10 kΩ.
- fNIRS Setup: Verify that all fNIRS optodes are in firm contact with the scalp. Check signal quality on the fNIRS acquisition software.
- Synchronization Check: Initiate a test recording to verify that event markers from the stimulus software are correctly recorded in both the EEG and fNIRS data streams.
4.2. Experimental Procedure The participant is seated comfortably in a chair approximately 1 meter from the monitor. The session structure is as follows:
A single trial within the block follows a structured timeline:
- Visual Cue (2 seconds): A directional arrow (pointing left or right) is displayed on the screen, instructing the participant which hand to imagine moving.
- Execution/MI Task (10 seconds): The screen displays a fixation cross. Following an auditory cue (a 200 ms beep), the participant performs kinesthetic motor imagery. They should imagine grasping with the cued hand at a rate of one grasp per second, without executing any physical movement [26].
- Inter-Trial Interval (15 seconds): A blank screen is shown, allowing the participant to rest and neural signals to return to baseline.
4.3. Data Acquisition Parameters Adhere to the following settings for consistent data quality:
- EEG Sampling Rate: 256 Hz or higher [26] [30].
- fNIRS Sampling Rate: ~11 Hz [26].
- fNIRS Wavelengths: Typically two wavelengths (e.g., 730 nm and 850 nm) to calculate concentration changes in oxygenated (HbO) and deoxygenated hemoglobin (HbR).
- Event Markers: Record the precise onset of the visual cue, execution phase, and end of trial in both data streams.
Data Processing and Analysis Workflow
Post-experiment, the synchronized data undergoes a multi-stage processing pipeline to extract meaningful features for BCI classification.
5.1. Pre-processing Steps
Table 3: Data Pre-processing Protocols
| Modality | Processing Step | Protocol Details & Parameters |
|---|---|---|
| EEG | Bandpass Filtering | Apply a 0.5-40 Hz filter to isolate relevant neural oscillations (e.g., Mu/Beta rhythms). |
| EEG | Artifact Removal | Use algorithms like Independent Component Analysis (ICA) to identify and remove components associated with eye blinks, eye movements, and muscle activity. |
| EEG | Epoching | Segment data into trials time-locked to the onset of the motor imagery cue (e.g., -2 to 15 seconds). |
| fNIRS | Optical Density Conversion | Convert raw light intensity signals to optical density. |
| fNIRS | Hemodynamic Response Calculation | Use the Modified Beer-Lambert Law to convert optical densities into concentration changes of HbO and HbR. |
| fNIRS | Bandpass Filtering | Apply a 0.01-0.2 Hz filter to remove physiological noise (e.g., cardiac cycle ~1 Hz, respiration ~0.3 Hz). |
| fNIRS | Epoching | Segment HbO/HbR data into trials aligned with the MI cue. |
5.2. Feature Extraction and Fusion
- EEG Features: Calculate the power spectral density in specific frequency bands (e.g., Mu 8-12 Hz, Beta 13-30 Hz) over the sensorimotor cortex during the task execution period [30].
- fNIRS Features: Extract the mean, slope, or peak value of the HbO signal during the task period, as HbO typically shows a more pronounced response during cortical activation [30].
- Data Fusion: Concatenate the selected EEG and fNIRS features into a single, high-dimensional feature vector for each trial. This hybrid feature set leverages the temporal precision of EEG and the spatial specificity of fNIRS, which has been shown to enhance classification accuracy by 5-10% compared to unimodal systems [26] [30].
Step-by-Step Guide to Simultaneous Data Acquisition and Signal Pre-processing
Simultaneous Electroencephalography (EEG) and functional Near-Infrared Spectroscopy (fNIRS) offer a powerful, multimodal approach for non-invasive brain imaging. By combining EEG's millisecond-level temporal resolution with fNIRS's superior spatial localization and hemodynamic monitoring, researchers can obtain a comprehensive view of brain activity [19] [30] [32]. This integrated methodology is particularly valuable for developing robust Brain-Computer Interfaces (BCIs), as it helps overcome the limitations inherent in using either modality alone, such as EEG's susceptibility to electrical noise and motion artifacts, and fNIRS's inherent physiological delay [30]. This protocol provides a detailed guide for the simultaneous acquisition and pre-processing of EEG and fNIRS data, framed within BCI research applications.
Hardware Setup and Data Acquisition
Equipment Configuration
The initial phase involves the physical integration of EEG and fNIRS systems. Careful configuration is essential to minimize interference and ensure temporal synchronization.
- Synchronization: A hardware trigger signal must be established between the EEG amplifier and the fNIRS system. This trigger, typically sent at the onset of each experimental trial or block, is crucial for aligning the two data streams during subsequent analysis. The fNIRS system's computer can often generate this signal via a parallel port or a dedicated digital I/O card.
- Electrode and Optode Placement: Position EEG electrodes and fNIRS optodes on the scalp according to the international 10-10 or 10-20 systems. Use custom caps that integrate holders for both to ensure stable positioning and minimize cross-talk. Prioritize the brain regions of interest for the specific BCI paradigm (e.g., motor cortex for motor imagery, prefrontal cortex for cognitive tasks). The distance between fNIRS source and detector optodes should generally be 3-4 cm to achieve a cortical penetration depth of approximately 2 cm [19] [30].
- Impedance and Signal Quality Check: For EEG, ensure that electrode-scalp impedances are reduced to below 10 kΩ, often requiring skin preparation and the application of conductive gel or paste [30]. For fNIRS, verify that optode-scalp coupling is sufficient by checking signal intensity levels and rejecting channels with poor light transmission.
Experimental Protocol for Data Acquisition
A standardized experimental protocol is critical for collecting high-quality, reproducible data. The following workflow outlines a typical session for a semantic decoding or mental imagery BCI paradigm, based on established research [19].
Table 1: Key Research Reagent Solutions and Materials
| Item | Function & Specification |
|---|---|
| EEG Amplifier System | Records electrical brain activity from the scalp. Requires high sampling rate (≥500 Hz) and synchronization input. |
| fNIRS System | Measures hemodynamic responses (HbO/HbR) using near-infrared light. Must support external triggering. |
| Integrated EEG/fNIRS Cap | Head cap with pre-configured layouts holding both EEG electrodes and fNIRS optodes for co-localized measurement. |
| EEG Electrodes & Gel | Ag/AgCl electrodes with conductive gel or paste are used to maintain signal fidelity and reduce impedance. |
| fNIRS Optodes | Sources emit NIR light; detectors measure light intensity after tissue penetration. |
| Stimulus Presentation Software | Software (e.g., PsychoPy, E-Prime) to present cues and send synchronization triggers. |
Signal Pre-processing Workflow
Raw, simultaneously acquired data requires modality-specific pre-processing to remove artifacts and noise before integrated analysis.
EEG Signal Pre-processing
EEG signals are weak and susceptible to various artifacts. The goal is to isolate neural activity from noise.
Core Steps:
- Downsampling: Reduce the sampling rate to decrease data volume and computational load while retaining sufficient information for analysis [32].
- Filtering: Apply band-pass filters (e.g., 0.5-45 Hz) to remove slow drifts and high-frequency noise, including line noise (e.g., 50/60 Hz notch filter) [32].
- Bad Channel Removal: Identify and interpolate channels with consistently poor signal quality.
- Artifact Removal: Use advanced algorithms like Independent Component Analysis (ICA) to separate and remove artifacts stemming from eye blinks, eye movements, and muscle activity [32].
- Epoching: Segment the continuous data into trials time-locked to the event triggers (e.g., from -1 s pre-stimulus to +5 s post-stimulus).
- Baseline Correction: Remove the mean signal from the pre-stimulus period from each epoch.
fNIRS Signal Pre-processing
fNIRS signals reflect hemodynamic changes and are contaminated by physiological noise and motion artifacts.
Core Steps:
- Optical Density Conversion: Convert raw light intensity measurements to optical density (OD) changes.
- Channel Pruning: Exclude channels with insufficient light intensity or extreme signal variance.
- Motion Artifact Correction: Identify and correct for signal spikes caused by participant movement using algorithms (e.g., wavelet-based, tPCA, or correlation-based methods).
- Band-Pass Filtering: Apply a filter (e.g., 0.01 - 0.2 Hz) to remove physiological noise such as cardiac (∼1 Hz) and respiratory (∼0.3 Hz) cycles, as well as very slow drifts.
- Hemoglobin Concentration Calculation: Use the Modified Beer-Lambert Law to convert the filtered OD changes into concentration changes for oxygenated (HbO) and deoxygenated (HbR) hemoglobin.
The following diagram illustrates the parallel pre-processing pipelines for both modalities and their point of integration.
Table 2: Standard Pre-processing Parameters for BCI Paradigms
| Processing Step | EEG | fNIRS |
|---|---|---|
| Sampling Rate | 500-1000 Hz (Acquisition) | 10-20 Hz (Acquisition) |
| Downsampling | 250 Hz | Not typically required |
| Filtering | Band-pass: 0.5-45 Hz; Notch: 50/60 Hz | Band-pass: 0.01-0.2 Hz |
| Key Artifact Removal | ICA for ocular & muscle artifacts | Wavelet or tPCA for motion artifacts |
| Epoching Baseline | -1.0 to 0.0 s pre-stimulus | -2.0 to 0.0 s pre-stimulus |
Integration and Final Considerations
After pre-processing, the epoched and cleaned EEG and fNIRS data are ready for integrated analysis. The high-temporal-resolution EEG features (e.g., Event-Related Potentials (ERPs), power in specific frequency bands) can be combined with the slower, hemodynamic fNIRS features (e.g., HbO/HbR slopes or means) to train machine learning or deep learning models for BCI control [19] [33] [32]. This hybrid approach can lead to more accurate and robust neural decoding than single-modality systems. Ensuring meticulous execution of both the acquisition and pre-processing phases, as detailed in this guide, is fundamental to the success of subsequent analysis and the overall BCI application.
Electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS) have emerged as prominent non-invasive neuroimaging techniques for brain-computer interface (BCI) research. Their integration creates a hybrid system that combines complementary characteristics - EEG provides millisecond-level temporal resolution of electrophysiological activity, while fNIRS offers superior spatial localization of hemodynamic responses with better resistance to motion artifacts [34] [4]. This complementary nature enables more comprehensive brain activity monitoring, particularly valuable for investigating complex cognitive processes and developing more robust BCI applications.
The effective integration of these modalities presents significant signal processing challenges. Multimodal fusion strategies have evolved to address these challenges through three primary approaches: data-level, feature-level, and decision-level fusion [4] [35]. Data-level fusion involves combining raw or preprocessed signals before feature extraction, preserving original information but requiring sophisticated synchronization and increasing computational complexity. Feature-level fusion merges extracted features from each modality before classification, effectively reducing data dimensionality while enhancing discriminative information. Decision-level fusion combines outputs from separate classifiers for each modality, offering robustness against modality-specific noise but potentially losing interactive information [36] [4].
This article provides a comprehensive technical resource detailing advanced methodologies for extracting and fusing EEG and fNIRS features, supported by experimental protocols and performance comparisons relevant to BCI researchers and developers.
Feature Extraction Techniques for EEG and fNIRS
EEG Feature Extraction Methods
EEG signal analysis focuses on capturing event-related changes in time, frequency, and spatial domains. Common Spatial Patterns (CSP) remains a widely used algorithm that identifies spatial filters that maximize variance for one class while minimizing it for another, particularly effective for motor imagery tasks [36]. For enhanced performance across subjects, the Filter Bank CSP (FBCSP) extends this approach by decomposing EEG signals into multiple frequency bands before applying CSP, addressing the variability in event-related synchronization/desynchronization characteristics across individuals [36].
Time-frequency analysis through Event-Related Spectral Perturbation (ERSP) characterizes power spectrum changes during tasks compared to baseline, enabling identification of frequency-specific neural oscillatory activity [36]. Deep learning approaches have demonstrated capability to automatically learn optimal feature representations from raw or minimally processed EEG data, potentially bypassing limitations of handcrafted features [4] [35].
fNIRS Feature Extraction Methods
fNIRS feature extraction primarily targets hemodynamic responses measured through oxygenated (HbO) and deoxygenated hemoglobin (HbR) concentration changes. Temporal statistical features including mean, maximum, slope, skewness, and kurtosis of hemoglobin concentration trajectories provide compact representations of hemodynamic response shapes [36]. The Modified CSP (MCSP) algorithm has been adapted for fNIRS signals, effectively extracting discriminative spatial patterns from hemodynamic responses [36].
Table 1: Performance Comparison of Fusion Strategies in BCI Applications
| Fusion Strategy | Average Accuracy (%) | Advantages | Limitations | Key Applications |
|---|---|---|---|---|
| Data-Level Fusion | Information not available | Preserves original signal information; Enables modeling of neurovascular coupling | High computational load; Sensitive to noise and artifacts; Requires precise temporal alignment | Neurovascular coupling studies; Source decomposition analysis [34] |
| Feature-Level Fusion | 77.53-88.33% [36] [4] | Reduces dimensionality; Enhances discriminative power; Flexible feature selection | Risk of feature redundancy; Requires careful normalization; Dependent on feature quality | Motor Imagery [36]; Mental Arithmetic [4]; Mental stress detection [37] |
| Decision-Level Fusion | 77.6% [4] | Robust to modality-specific noise; Enables heterogeneous processing; Modular implementation | Potential loss of inter-modal dynamics; Requires separate classifiers | Mental stress detection [37] [4]; Compact hBCI systems [4] |
Multimodal Fusion Frameworks
Data-Level Fusion
Data-level fusion, also called early fusion, involves combining raw or minimally processed signals from EEG and fNIRS before feature extraction. This approach aims to exploit potential couplings between electrophysiological and hemodynamic responses at their most fundamental level [34]. The primary advantage lies in preserving the original signal information, potentially enabling more sophisticated models of neurovascular coupling mechanisms underlying brain activity.
Implementation typically requires temporal alignment of signals with different sampling rates (EEG: typically 256 Hz or higher; fNIRS: typically 10-20 Hz) through resampling or interpolation techniques [26]. Advanced source decomposition techniques like joint independent component analysis (jICA) can be applied to the fused data to identify latent components representing shared neural processes [34] [37]. However, this approach demands substantial computational resources and careful artifact removal to prevent noise propagation through the analysis pipeline.
Feature-Level Fusion
Feature-level fusion represents the most widely adopted approach in EEG-fNIRS BCI research, combining extracted features from each modality before classification [36] [4]. This strategy effectively reduces data dimensionality while leveraging complementary information from both modalities.
Common techniques include feature concatenation, where feature vectors from EEG and fNIRS are combined into a single comprehensive vector [37] [36]. For two-class motor imagery tasks, this approach has achieved accuracies of 88.33% when combining EEG with HbO and HbR features [36]. Advanced methods like multi-resolution singular value decomposition (MSVD) enable system-level fusion by decomposing signals into approximation and detail coefficients, providing a structured framework for integrating complementary information [37].
Feature selection algorithms play a crucial role in optimizing feature-level fusion. The combination of Relief and minimum redundancy maximum relevance (mRMR) algorithms has demonstrated effectiveness in identifying optimal feature subsets, significantly reducing feature dimensionality while improving classification performance [36]. Deep learning approaches have also been employed for automated feature fusion, with tensor fusion and p-th order polynomial fusion achieving 77.53% and 90.19% accuracy for motor imagery and mental arithmetic tasks, respectively [4].
Decision-Level Fusion
Decision-level fusion, or late fusion, combines outputs from separate classifiers trained on modality-specific features [4] [35]. This approach maintains modality independence during processing while leveraging their complementary nature at the decision stage, offering inherent robustness against modality-specific artifacts and noise.
Implementation typically involves training separate classifiers for EEG and fNIRS features, then combining their probabilistic outputs or decisions through various strategies. Weighted averaging of classifier outputs based on modality reliability has demonstrated significant performance improvements, with one study reporting approximately 5% average accuracy improvement across subjects compared to single-modality approaches [38]. Meta-classifier frameworks train a secondary classifier on the outputs of modality-specific classifiers, effectively learning optimal integration strategies [38].
Recent advances incorporate cross-modal attention mechanisms that dynamically weight the importance of each modality based on task context. The MBC-ATT framework employs independent branch structures to process EEG and fNIRS signals separately, with an attention mechanism that selectively emphasizes relevant features across modalities, demonstrating superior performance for cognitive task classification [35].
Experimental Protocols and Applications
Motor Imagery Paradigm
Objective: To decode motor intention through imagined movements for BCI control and neurorehabilitation applications, particularly in stroke and intracerebral hemorrhage patients [26] [36].
Participants: Protocol validated with both healthy subjects (n=17-18) and patients with intracerebral hemorrhage (n=20) [26] [36].
Experimental Setup:
- Participants seated comfortably 25cm from display monitor
- 32-channel EEG system (256Hz sampling) synchronized with fNIRS system (11Hz sampling)
- 32 optical sources and 30 photodetectors arranged in optimized topography (90 fNIRS channels) [26]
Procedure:
- Baseline recording: 1-minute eyes-closed followed by 1-minute eyes-open states
- Trial structure:
- Visual cue presentation (2s): Directional arrow (left/right) indicating required motor imagery
- Execution phase (10s): Participants perform kinesthetic motor imagery of grasping movement with corresponding hand at 1Hz rate
- Inter-trial interval (15s): Blank screen for rest [26]
- Session design: Minimum 2 sessions with 30 trials each (15 left/right hand imagery)
- Enhancement strategy: Incorporate grip strength calibration with dynamometer before data acquisition to enhance motor imagery vividness [26]
Data Analysis:
- Extract CSP features from EEG signals across multiple frequency bands
- Calculate temporal statistical features and MCSP features from fNIRS HbO and HbR signals
- Apply Relief-mRMR feature selection to identify optimal feature subset
- Implement feature-level fusion with SVM classification [36]
Cognitive Task Paradigm
Objective: To investigate brain activity during higher cognitive functions including working memory, language processing, and mental arithmetic for advanced BCI applications [19] [35].
Participants: Studies conducted with 26 healthy right-handed adults (ages 17-33) [35].
Experimental Tasks:
- n-back task (working memory):
- 0-back, 2-back, and 3-back conditions
- Participants determine if current number matches number presented 2 or 3 trials earlier
- Task structure: 2s instruction → 40s task period → 20s rest [35]
Word Generation task (language processing):
- Participants generate words starting with specific letter during task period
- Task structure: 2s task prompt → 10s word generation → 13-15s rest [35]
Mental Arithmetic task:
- Participants perform arithmetic calculations mentally without verbalization or movement
- Typically involves serial subtraction or multiplication [4]
Data Analysis:
- For EEG: Extract time-frequency features (theta, alpha power) and ERP components (P300)
- For fNIRS: Analyze HbO and HbR concentration changes in prefrontal cortex
- Implement cross-modal attention fusion (MBC-ATT) for classification [35]
Table 2: The Scientist's Toolkit: Essential Research Reagents and Materials
| Category | Item | Specification/Model | Function/Application |
|---|---|---|---|
| Acquisition Hardware | EEG Amplifier | g.HIamp amplifier (g.tec) | High-quality EEG signal acquisition with 256Hz sampling [26] |
| fNIRS System | NirScan (Danyang Huichuang) | Continuous-wave hemodynamic measurement with 11Hz sampling [26] | |
| Hybrid Cap | Custom design with 32 EEG electrodes, 32 sources, 30 detectors | Simultaneous positioning of EEG and fNIRS optodes [26] | |
| Software & Analysis | Stimulus Presentation | E-Prime 3.0 | Precise experimental paradigm control and event marker generation [26] |
| EEG Analysis | EEGLab | Preprocessing, ERP analysis, and time-frequency decomposition [39] | |
| Feature Extraction | Custom MATLAB/Python scripts | Implementation of CSP, FBCSP, and statistical feature extraction [36] | |
| Classification | SVM, CNN-LSTM, MBC-ATT | Multimodal classification and fusion [36] [35] | |
| Experimental Materials | Response Device | Numeric keypad | Participant responses during cognitive tasks [35] |
| Calibration Tools | Dynamometer, stress ball | Enhancing motor imagery vividness through tactile reinforcement [26] |
Performance Analysis and Future Directions
Comparative Performance Evaluation
Research consistently demonstrates that multimodal approaches outperform single-modality systems across various BCI paradigms. For motor imagery tasks, feature-level fusion of EEG with fNIRS HbO and HbR features has achieved 88.33% classification accuracy, significantly exceeding EEG-only performance (84.28%) [36]. In mental arithmetic tasks, advanced fusion frameworks combining multi-domain features with multi-level progressive learning have reached remarkable 98.42% accuracy [4].
The complementary nature of EEG and fNIRS is evidenced by their differential sensitivity to various cognitive states. While EEG excellently captures rapid neural dynamics during task onset, fNIRS provides sustained monitoring of hemodynamic responses during prolonged cognitive engagement [33]. This temporal complementarity enables more comprehensive brain state monitoring across different timescales.
Emerging Trends and Future Directions
Deep learning architectures specifically designed for multimodal fusion represent a promising research direction. Models incorporating cross-modal attention mechanisms dynamically weight the importance of each modality based on task context, significantly enhancing fusion effectiveness [35]. The MBC-ATT framework demonstrates how modality-guided attention can selectively integrate EEG and fNIRS features, improving decoding accuracy for cognitive tasks.
Clinical translation remains a crucial frontier, with recent datasets specifically addressing pathological populations such as intracerebral hemorrhage patients [26]. Developing personalized fusion models that adapt to individual neurovascular coupling characteristics and pathological conditions will be essential for real-world clinical applications.
Hardware advancements enabling higher-density arrangements with improved signal quality will further enhance spatial resolution and depth sensitivity. Combined with real-time processing algorithms, these improvements will support more sophisticated fusion approaches in practical BCI systems for communication, rehabilitation, and cognitive enhancement.
Machine Learning and Deep Learning Applications for Multimodal Classification
The integration of electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS) has emerged as a transformative approach in brain-computer interface (BCI) research, offering a synergistic combination of temporal and spatial resolution for decoding neural activity. Multimodal classification, powered by machine learning (ML) and deep learning (DL), is pivotal for translating the complementary information from these modalities into robust BCI systems [2] [40]. EEG provides millisecond-level temporal resolution for capturing rapid neuronal dynamics, while fNIRS offers superior spatial localization and resistance to motion artifacts by measuring hemodynamic responses associated with neural activity [26] [30]. This complementary relationship addresses the inherent limitations of each unimodal approach, enabling enhanced classification accuracy and more reliable systems for applications such as motor imagery (MI) decoding and cognitive state monitoring [40] [30]. This document provides detailed application notes and experimental protocols for multimodal EEG-fNIRS classification, framed within the context of a simultaneous setup for BCI research.
Available Multimodal EEG-fNIRS Datasets
Publicly available datasets are essential for developing and benchmarking ML/DL models. The table below summarizes key datasets used in contemporary research.
Table 1: Publicly Available Multimodal EEG-fNIRS Datasets for Classification
| Dataset Name | Subjects (Healthy/Patients) | Recorded Tasks | Key Modality Specifications | Primary Research Use |
|---|---|---|---|---|
| HEFMI-ICH [26] | 17 Normal, 20 Intracerebral Hemorrhage (ICH) patients | Left/Right Hand Motor Imagery | 32 EEG channels (256 Hz), 90 fNIRS channels (11 Hz) | ICH Rehabilitation, Patient-Specific Model Development |
| Shin et al. (2018) [40] | 26 Healthy | n-back, Discrimination/Selection Response, Word Generation | 30 EEG channels (1000 Hz), 36 fNIRS channels (12.5 Hz) | Cognitive State Decoding (Working Memory, Language) |
| Simultaneous EEG-fNIRS for Structure-Function Analysis [27] | 18 Healthy | Resting State, Left/Right Hand Motor Imagery | 30 EEG channels (1000 Hz), 36 fNIRS channels (12.5 Hz) | Investigating Structure-Function Relationships, Network Analysis |
Machine Learning and Deep Learning Frameworks for Classification
Multimodal classification frameworks can be broadly categorized by their fusion strategy: early fusion (combining raw data or low-level features), late fusion (combining high-level features or decisions), and hybrid approaches that use advanced mechanisms to model cross-modal interactions.
Table 2: Classification Algorithms and Performance for EEG-fNIRS BCI
| Model Category | Specific Model/Architecture | Fusion Strategy & Key Innovation | Reported Application & Performance |
|---|---|---|---|
| Deep Learning (Hybrid) | Multi-Branch CNN with Attention (MBC-ATT) [40] | Late fusion with a cross-modal attention mechanism. Dynamically weights the importance of features from each modality. | Cognitive Task (n-back, WG) Classification. Outperformed conventional approaches. |
| Deep Learning (Hybrid) | RP-based time-distributed CNN-LSTM [40] | Late fusion. Uses Recurrence Plots (RP) to represent temporal dynamics for CNN, with LSTM capturing long-term dependencies. | Integrated classification of EEG and fNIRS signals in hybrid BCI. |
| Deep Learning (Hybrid) | STFT + DenseNet [40] | Intermediate fusion. Converts EEG to time-frequency images via STFT and integrates with fNIRS frequency features using DenseNet. | Enhanced multimodal representation and classification performance. |
| Traditional ML | Handcrafted Features + Traditional Classifiers [40] | Early or late fusion. Relies on manually extracted features (e.g., band power for EEG, HbO/HbR concentration for fNIRS) fed into classifiers like SVM or LDA. | Classifying multi-level brain load. Heavily relies on preprocessing and feature engineering. |
| Unified Framework | Feature-level fusion with adaptive weighting [26] | Early/Intermediate fusion. Combines engineered features from both modalities with adaptive mechanisms to enhance synergy. | Neural signal classification in both healthy subjects and ICH patients. |
Detailed Framework: MBC-ATT with Cross-Modal Attention
The MBC-ATT framework represents a significant advancement in late fusion strategies [40]. Its architecture and workflow are detailed below.
Experimental Protocols
This section outlines standardized protocols for data acquisition, preprocessing, and model training tailored for multimodal EEG-fNIRS classification.
Simultaneous Data Acquisition Protocol
Objective: To acquire high-quality, temporally synchronized EEG and fNIRS data during a motor imagery (MI) paradigm.
Materials: Refer to "The Scientist's Toolkit" (Section 6) for essential equipment.
Procedure:
Participant Preparation & Consent:
- Recruit participants following ethical guidelines (e.g., approved by an institutional review board). Obtain written informed consent [26].
- Prepare the scalp according to standard procedures (e.g., cleansing for EEG electrode placement). Measure head circumference to select an appropriately sized cap [26].
Equipment Setup & Synchronization:
- Use a custom-designed hybrid EEG-fNIRS cap that integrates both electrodes and optodes in a predefined topography [26] [2].
- For EEG, place electrodes according to the international 10-20 or 10-5 system [27] [40]. For fNIRS, arrange sources and detectors to cover regions of interest (e.g., motor cortex) with a standard source-detector separation (e.g., 3 cm) [26].
- Ensure temporal synchronization between the EEG and fNIRS systems. This can be achieved by using a unified processor [2] or by transmitting event markers from the stimulus presentation software (e.g., E-Prime) to both recording systems simultaneously [26].
Motor Imagery Paradigm Execution:
- Seat the participant in a comfortable chair approximately 1-1.2 meters from a monitor [26] [40].
- The MI paradigm should be structured as follows for each trial [26]:
- Visual Cue (2 s): A directional arrow (left or right) is displayed, instructing the participant which hand to imagine moving.
- Execution/MI Phase (10 s): A fixation cross is displayed. The participant performs kinesthetic motor imagery of grasping with the cued hand at a rate of one imagined grasp per second.
- Inter-Trial Interval (15 s): A blank screen is shown for rest.
- Conduct multiple sessions (e.g., 2+), each containing a sufficient number of trials (e.g., 15-30 per class), with rest intervals between sessions to mitigate fatigue [26].
Data Preprocessing Pipeline Protocol
Objective: To prepare raw EEG and fNIRS signals for feature extraction and model input.
Procedure:
EEG Preprocessing:
- Filtering: Apply a bandpass filter (e.g., 0.5-40 Hz) to remove low-frequency drift and high-frequency noise [40].
- Re-referencing: Re-reference the data to a common average or mastoid reference.
- Artifact Removal: Identify and remove artifacts from eye movements (EOG) and muscle activity (EMG) using techniques like Independent Component Analysis (ICA) or regression-based methods [40].
fNIRS Preprocessing:
- Signal Quality Check: Calculate the Scalp-Coupling Index (SCI) to identify and exclude low-quality channels [27].
- Optical Density Conversion: Convert raw light intensity signals to optical densities.
- Filtering: Bandpass filter (e.g., 0.01-0.1 Hz for resting state; 0.02-0.08 Hz for task-based) to isolate the hemodynamic response and reduce physiological noise (heart rate, respiration) [27].
- Motion Artifact Correction: Use algorithms like PCA or targeted motion correction (e.g., based on the GVTD metric) to remove motion artifacts [27].
- Hemoglobin Concentration Calculation: Convert preprocessed optical densities to changes in oxygenated (HbO) and deoxygenated (HbR) hemoglobin concentrations using the Modified Beer-Lambert Law [30].
Epoching & Labeling:
- Segment the continuous preprocessed data into epochs/time windows relative to task onset (e.g., 0-10 s after the MI cue) [27].
- Assign the correct class label (e.g., "Left Hand MI", "Right Hand MI") to each epoch.
Model Training & Evaluation Protocol
Objective: To train and validate a multimodal classification model using the preprocessed data.
Procedure:
Feature Extraction (for Traditional ML):
Data Splitting:
- Partition the epoched data into training, validation, and test sets. Use a subject-independent (leave-one-subject-out) cross-validation strategy to rigorously evaluate model generalizability [40].
Model Implementation & Training:
- For DL models (e.g., MBC-ATT), implement the architecture in a framework like PyTorch or TensorFlow.
- Train the model using the training set, monitoring performance on the validation set to avoid overfitting. Use optimizers like Adam and a categorical cross-entropy loss function.
Evaluation & Reporting:
Signaling Pathways and Logical Workflows
The following diagram illustrates the logical relationship between neural activity, the signals measured by EEG and fNIRS, and the subsequent processing for BCI classification.
The Scientist's Toolkit: Research Reagent Solutions
This table details the essential materials, software, and analytical "reagents" required for conducting multimodal EEG-fNIRS classification experiments.
Table 3: Essential Research Reagents and Materials for EEG-fNIRS BCI Research
| Item Category | Specific Examples & Specifications | Primary Function in Research Workflow |
|---|---|---|
| Acquisition Hardware | Hybrid EEG-fNIRS Cap (e.g., integrated 32 EEG electrodes + 90 fNIRS channels) [26] | Provides the physical interface for simultaneous signal acquisition; ensures proper scalp contact and co-registration of modalities. |
| Signal Amplifiers & Systems | g.HIamp EEG Amplifier (g.tec); NirScan fNIRS System (Danyang Huichuang) [26] | Amplifies and digitizes weak analog biological signals from the scalp for subsequent processing. |
| Stimulus Presentation Software | E-Prime 3.0 (Psychology Software Tools) [26] | Presents experimental paradigms, records participant responses, and sends synchronization triggers to recording hardware. |
| Data Preprocessing Tools | MNE-Python, Brainstorm, EEGLAB, Homer2, NIRS-KIT | Provides standardized pipelines for filtering, artifact removal, epoching, and converting raw signals into analyzable data. |
| Feature Extraction Libraries | Python (scikit-learn, MNE), MATLAB (Signal Processing Toolbox) | Automates the computation of relevant features (e.g., CSP, band power, HbO statistics) from preprocessed data. |
| Machine Learning Frameworks | Scikit-learn (for SVM, LDA, etc.) [40] | Offers implementations of traditional machine learning models for classification with handcrafted features. |
| Deep Learning Frameworks | PyTorch, TensorFlow/Keras [40] | Provides flexible environments for building, training, and evaluating complex deep learning models like MBC-ATT and CNN-LSTMs. |
| Validation & Metrics | Subject-Independent Cross-Validation, scikit-learn metrics | Ensures rigorous evaluation of model generalizability and provides standard performance measures (Accuracy, F1-Score). |
The development of Brain-Computer Interfaces (BCIs) has been significantly advanced by multimodal neuroimaging approaches. The simultaneous acquisition of Electroencephalography (EEG) and functional Near-Infrared Spectroscopy (fNIRS) represents a particularly powerful combination for decoding brain states, leveraging the complementary strengths of both modalities. EEG provides excellent temporal resolution at the millisecond level, capturing rapid neuronal activation patterns, while fNIRS tracks slower hemodynamic responses with superior spatial localization, offering insights into metabolic demands of neural activity [5] [1]. This integration is especially valuable for studying neurovascular coupling - the fundamental relationship between electrical brain activity and subsequent hemodynamic changes [1]. The following application notes and protocols detail how this hybrid approach is being successfully implemented across three distinct domains: motor imagery, mental arithmetic, and clinical diagnostics, providing researchers with practical frameworks for implementing these methodologies in BCI research.
Application Notes & Experimental Protocols
Motor Imagery for Rehabilitation BCIs
Application Note: Motor Imagery (MI)-based BCIs have emerged as a transformative approach for post-stroke rehabilitation, leveraging neuroplasticity to facilitate motor network reorganization through closed-loop feedback mechanisms [26]. The hybrid EEG-fNIRS approach capitalizes on their spatiotemporal synergy: EEG captures rapid neuronal activation patterns during MI tasks, while fNIRS tracks slower hemodynamic changes associated with cortical reorganization [26]. This combination has demonstrated 5%-10% improvement in classification accuracy compared to unimodal systems in healthy subjects, with recent work extending these benefits to clinical populations such as intracerebral hemorrhage (ICH) patients [26].
Table 1: Key Datasets for Motor Imagery BCI Research
| Dataset Name | Modality | Participants | MI Paradigm | Key Features | Reference |
|---|---|---|---|---|---|
| HEFMI-ICH | EEG-fNIRS | 17 healthy, 20 ICH patients | Left/right hand kinesthetic MI | First hybrid dataset for ICH rehabilitation; provides raw and preprocessed data | [26] |
| Yi et al. Multimodal Dataset | EEG-fNIRS | Multiple subjects | Upper limb MI without real motion | Comprehensive open dataset for neurovascular coupling during mental simulation | [41] |
| TU-Berlin-A | EEG-fNIRS | Not specified | Motor imagery | Publicly available for classification algorithm development | [17] |
Experimental Protocol:
Participant Preparation and Training:
- Recruit participants (healthy controls or patients with motor deficits) and obtain informed consent.
- For patients with ICH, conduct clinical assessments (Fugl-Meyer Assessment for Upper Extremities, Modified Barthel Index, modified Rankin Scale) [26].
- Enhance MI vividness through a grip strength calibration procedure using a dynamometer and stress ball to reinforce tactile and force-related aspects of movement [26].
Data Acquisition:
- Equipment: Use a synchronized EEG-fNIRS system. Example: g.HIamp amplifier (EEG) and NirScan system (fNIRS) with a custom hybrid cap integrating 32 EEG electrodes and 62 fNIRS optodes (32 sources, 30 detectors) providing 90 measurement channels [26].
- Setup: Participants sit 25 cm from a display monitor. Record 1-minute eyes-closed and 1-minute eyes-open baseline signals [26].
- Paradigm: Implement an event-based design. Each trial consists of:
- Conduct at least 2 sessions (60 trials total), with adjustable breaks between sessions to mitigate fatigue [26].
Signal Processing and Analysis:
- Preprocessing:
- fNIRS: Convert raw intensity to optical density, then to hemoglobin concentration (HbO, HbR) using the Modified Beer-Lambert Law [42]. Apply band-pass filter (0.05-0.7 Hz) to remove cardiac pulsation and low-frequency drift [42].
- EEG: Apply appropriate filters and artifact correction methods. Note: The choice of preprocessing steps (filtering, artifact correction) significantly impacts subsequent decoding performance [43].
- Data Analysis: Extract spatiotemporal features from EEG and hemodynamic features from fNIRS. Employ advanced fusion methods such as deep learning with evidence theory for classification, which has achieved accuracies up to 83.26% for MI tasks [17].
- Preprocessing:
Figure 1: Experimental workflow for a hybrid EEG-fNIRS Motor Imagery BCI protocol.
Semantic Decoding and Mental Workload
Application Note: fNIRS demonstrates significant advantages for monitoring cognitive tasks in the prefrontal cortex, a region relatively free from hair coverage interference [5]. Semantic neural decoding aims to identify which semantic concepts an individual focuses on based on brain activity, enabling a new type of BCI that communicates conceptual meaning directly, bypassing character-by-character spelling used in current systems [19]. Simultaneous EEG-fNIRS recordings during semantic tasks (e.g., categorizing animals vs. tools) leverage fNIRS's sensitivity to hemodynamic changes in prefrontal regions combined with EEG's temporal resolution to track rapid neural dynamics during cognitive processing.
Table 2: Semantic Decoding Tasks and Modalities
| Mental Task | Instruction to Participant | Primary Neural Correlate | Modality Advantages |
|---|---|---|---|
| Silent Naming | Silently name the displayed object | Language processing networks | fNIRS: Prefrontal hemodynamics; EEG: Temporal dynamics of word retrieval |
| Visual Imagery | Visualize the object in your mind | Visual association cortex | Combined approach distinguishes category-specific patterns |
| Auditory Imagery | Imagine sounds the object makes | Auditory cortex | EEG captures temporal sound structure; fNIRS monitors sustained processing |
| Tactile Imagery | Imagine feeling of touching the object | Somatosensory cortex | Multisensory integration provides robust decoding features |
Experimental Protocol:
Stimuli and Paradigm:
- Stimuli Selection: Prepare images from distinct semantic categories (e.g., 18 animals and 18 tools). Use photographic images converted to grayscale and standardized [19].
- Task Design: Implement a block or event-related design. Present stimuli and instruct participants to perform specific mental tasks:
- Timing: Each task period should last 3-5 seconds, followed by adequate rest periods to allow hemodynamic response return to baseline [19].
Data Acquisition:
- Equipment: Use synchronized EEG-fNIRS systems with optodes and electrodes covering prefrontal and temporal regions.
- Participant Selection: Include native speakers for language-based tasks to minimize variability in neural representation of semantic concepts [19].
Signal Processing and Analysis:
- fNIRS Processing: Convert signals to HbO and HbR concentrations. Apply physiological noise removal (band-pass filter 0.01-0.2 Hz) to isolate task-related hemodynamic responses [44].
- EEG Processing: Preprocess to remove artifacts and extract event-related potentials or spectral features.
- Data Fusion: Employ machine learning classifiers (e.g., SVM, LDA) or deep learning models to distinguish between semantic categories based on combined features.
Figure 2: Signal processing pathway for hybrid EEG-fNIRS semantic decoding.
Clinical Diagnostics and Neurorehabilitation
Application Note: Hybrid EEG-fNIRS offers significant potential for clinical diagnostics and monitoring neurorehabilitation progress, particularly in stroke recovery. ICH patients present unique neurovascular coupling challenges where unimodal approaches may fail due to disrupted neurovascular pathways [26]. The combined approach allows clinicians to monitor both electrical and hemodynamic aspects of recovery, providing a more complete picture of brain reorganization. fNIRS is particularly valuable in clinical populations due to its tolerance for movement and suitability for bedside monitoring [1].
Experimental Protocol:
Patient Assessment and Paradigm Adaptation:
- Conduct comprehensive clinical assessments specific to the patient population (e.g., FMA-UE, MBI, mRS for stroke) [26].
- Adapt task paradigms to accommodate potential cognitive or motor limitations of patients.
- Simplify instructions and provide practice sessions to ensure task comprehension.
Data Acquisition in Clinical Settings:
- Use portable, quick-setup systems suitable for clinical environments.
- Ensure patient comfort during recording sessions, which may need to be shorter than research studies.
- Position optodes and electrodes to cover affected and unaffected hemispheres for comparison.
Clinical-Specific Processing and Analysis:
- Artifact Handling: Implement robust motion artifact correction techniques suitable for patient populations who may have difficulty remaining still [44].
- Asymmetry Analysis: Calculate inter-hemispheric asymmetry indices for both EEG and fNIRS features as potential biomarkers of recovery.
- Longitudinal Tracking: Monitor changes in neural and hemodynamic activity across multiple sessions to track neuroplasticity and rehabilitation progress.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials and Analytical Tools for EEG-fNIRS Research
| Category | Item | Specification/Function | Example Use Cases |
|---|---|---|---|
| Recording Equipment | Hybrid EEG-fNIRS Cap | Integrated electrodes and optodes with standardized positioning | Motor imagery studies, clinical patient monitoring [26] |
| fNIRS System (CW) | Continuous wave system with dual wavelengths (~690, ~830 nm) | Hemodynamic monitoring in cognitive tasks, bedside monitoring [5] [1] | |
| EEG Amplifier | High-input impedance, >250 Hz sampling rate | Capturing event-related potentials during cognitive tasks [26] | |
| Software & Analysis | MNE-Python | Open-source Python package for EEG/fNIRS processing | Preprocessing pipelines, feature extraction, visualization [42] |
| Preprocessing Tools | Band-pass filters, ICA, artifact correction algorithms | Removing physiological noise, motion artifacts [42] [44] | |
| Classification Algorithms | SVM, LDA, EEGNet, Deep Learning Fusion Models | Motor imagery classification, semantic decoding [17] [7] [43] | |
| Experimental Materials | Grip Strength Tools | Dynamometer, stress balls | Enhancing MI vividness through kinesthetic reinforcement [26] |
| Stimulus Presentation Software | E-Prime, PsychoPy, Presentation | Precise timing control for task paradigms [26] | |
| Validation Resources | Public Datasets | HEFMI-ICH, Yi et al. multimodal dataset | Algorithm validation, comparative studies [41] [26] |
The integration of EEG and fNIRS technologies presents a powerful multimodal framework for advancing BCI research across diverse applications. The protocols outlined for motor imagery, semantic decoding, and clinical diagnostics provide researchers with practical methodologies for implementing this hybrid approach. The complementary nature of electrical and hemodynamic signals offers a more comprehensive window into brain function than either modality alone, enabling improved classification accuracy and more nuanced understanding of brain states. As processing algorithms continue to evolve—particularly deep learning and evidence theory-based fusion methods—and standardized datasets become more available, the potential for transformative applications in both basic neuroscience and clinical practice continues to expand. Future directions should focus on refining real-time processing capabilities, enhancing spatial resolution through high-density arrays, and developing more adaptive classification frameworks that can accommodate individual variability in neural signatures.
Overcoming Practical Challenges: Signal Quality, Data Processing, and System Optimization
Simultaneous electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS) setups represent a powerful multimodal approach for brain-computer interface (BCI) research, combining EEG's excellent temporal resolution with fNIRS's good spatial specificity [18] [1]. However, the data acquired from these hybrid systems are frequently contaminated by several types of artifacts that can compromise data quality and interpretation. Motion artifacts, physiological noise, and signal crosstalk constitute the primary challenges that researchers must address to ensure the reliability of their findings. This application note provides detailed protocols and methodologies for identifying and mitigating these common artifacts, thereby enhancing the signal quality and validity of simultaneous EEG-fNIRS studies in BCI applications.
Motion Artifacts: Correction Protocols and Performance
Motion artifacts originate from subject movement, causing sudden shifts in signal baseline, high-frequency spikes, or slow drifts in both EEG and fNIRS data [45] [46]. These artifacts can severely distort the neural signals of interest, particularly in paradigms involving patient movement or lengthy recording sessions.
Wavelet-Based Motion Correction Techniques
Wavelet-based methods provide effective approaches for motion artifact correction in single-channel recordings. The Wavelet Packet Decomposition (WPD) and WPD with Canonical Correlation Analysis (WPD-CCA) techniques have demonstrated significant efficacy in reducing motion artifacts.
Table 1: Performance Comparison of Motion Artifact Correction Methods
| Method | Modality | Average ΔSNR (dB) | Average Artifact Reduction (%) | Key Parameters |
|---|---|---|---|---|
| WPD (db2) | EEG | 29.44 | 53.48 | Decomposition level: 4; Wavelet: db2 |
| WPD-CCA (db1) | EEG | 30.76 | 59.51 | Decomposition level: 4; Wavelet: db1 |
| WPD (fk4) | fNIRS | 16.11 | 26.40 | Decomposition level: 4; Wavelet: fk4 |
| WPD-CCA (db1) | fNIRS | 16.55 | 41.40 | Decomposition level: 4; Wavelet: db1 |
The WPD method decomposes signals into wavelet packet bases at multiple scales, allowing for selective reconstruction of neural components while excluding artifact-dominated components [45]. The WPD-CCA approach extends this by applying canonical correlation analysis to the decomposed signals to further separate neural activity from motion artifacts [45].
Experimental Protocol: WPD-CCA Motion Correction
- Signal Acquisition: Record single-channel EEG or fNIRS data at appropriate sampling rates (EEG: ≥250 Hz; fNIRS: ≥10 Hz).
- Wavelet Selection: Choose an appropriate wavelet family (Daubechies for EEG; Fejer-Korovkin for fNIRS).
- Signal Decomposition: Perform 4-level wavelet packet decomposition using the selected wavelet.
- Component Analysis: Apply CCA to identify and separate artifact components from neural signals.
- Signal Reconstruction: Reconstruct the corrected signal using artifact-free components.
- Validation: Calculate ΔSNR and percentage artifact reduction to verify performance.
Learning-Based Motion Artifact Processing
Recent advances in machine learning and deep learning have introduced powerful alternatives for motion artifact correction. These methods are particularly valuable for handling large fNIRS datasets and complex artifact patterns [46].
Table 2: Learning-Based Approaches for Motion Artifact Removal
| Method | Architecture | Application | Performance Metrics |
|---|---|---|---|
| Wavelet Regression ANN | Artificial Neural Network | fNIRS | Contrast-to-Noise Ratio (CNR) |
| Motion Artifact Classification | SVM, KNN, GBT | fNIRS Vigilance Detection | Classification Accuracy |
| U-Net HRF Reconstruction | Convolutional Neural Network | fNIRS | Mean Squared Error (MSE) |
| Denoising Auto-Encoder | Auto-Encoder Model | fNIRS | Signal-to-Noise Ratio (SNR) |
| sResFCNN with FIR Filter | Fully Connected Neural Network | fNIRS | ΔSNR, Artifact Reduction |
Experimental Protocol: Deep Learning Motion Correction
- Data Preparation: Create a training dataset with clean fNIRS signals and simulated motion artifacts.
- Network Selection: Choose an appropriate network architecture (e.g., U-Net for hemodynamic response reconstruction).
- Model Training: Train the network to reconstruct artifact-free signals from contaminated inputs.
- Model Validation: Test the trained model on experimental data with known artifacts.
- Performance Quantification: Evaluate using metrics such as ΔSNR, MSE, and classification accuracy.
Motion Artifact Correction Methods
Physiological Noise: Mitigation Strategies
Physiological noise in fNIRS data arises from cardiac activity (~1 Hz), respiration (~0.25 Hz), Mayer waves (~0.1 Hz), and other systemic physiological processes [47]. These noises share frequency components with the hemodynamic response, making their removal particularly challenging.
Adaptive Filtering for Physiological Noise
Recursive least-squares estimation (RLSE) with an exponential forgetting factor provides an effective approach for physiological noise removal in fNIRS data. This method models the measured signal as a linear combination of the expected hemodynamic response, short-separation measurement data, physiological noises, and baseline drift [47].
The physiological noise model incorporates three principal components:
- Cardiac activity: Modeled as sinusoidal function at ~1 Hz
- Respiration: Modeled as sinusoidal function at ~0.25 Hz
- Mayer waves: Modeled as sinusoidal function at ~0.1 Hz
Experimental Protocol: RLSE Physiological Noise Removal
- Signal Model Definition: Formulate the linear regression model: y(t) = a₁u(t) + a₂Δu(t) + a₃Δ²u(t) + a₄ySS(t) + Σ[bmsin(2πfmt)] + b₀ + ε(t) where u(t) is the expected HR, ySS(t) is short-separation data, and the sinusoidal components represent physiological noises.
Short-Separation Channel Integration: Place optodes with separation <1 cm to capture superficial layer noises.
Parameter Estimation: Apply RLSE with exponential forgetting to estimate model parameters.
Noise Removal: Subtract the estimated physiological noise components from the measured signal.
Validation: Quantify performance using contrast-to-noise ratio improvements.
PCA-Based Denoising with Short-Separation Channels
For whole-head fNIRS montages, an automated denoising method incorporating principal component analysis (PCA) and general linear model (GLM) can effectively identify and remove globally uniform superficial components [48].
Experimental Protocol: PCA-Based Denoising
- Montage Setup: Implement a high-density montage with both long-separation (>3 cm) and short-separation (~1.5 cm) channels.
Data Collection: Acquire simultaneous recordings from all channels during task performance.
Component Analysis: Apply PCA to identify global superficial components.
Regression Modeling: Use GLM to regress out identified noise components.
Topographic Mapping: Generate cleaned whole-head topography of fNIRS activation.
This approach has demonstrated the ability to reveal focal activations concurrently in primary motor and visual areas after denoising [48].
Physiological Noise Removal Approaches
Signal Crosstalk: Evaluation and Reduction
Crosstalk in multi-channel, multi-parameter NIRS systems refers to unwanted signal interference between adjacent channels or parameters, which can significantly reduce measurement accuracy and reliability [49].
Crosstalk Evaluation Methods
A comprehensive set of test methods has been developed to evaluate crosstalk in NIRS instruments:
Human Blood Model Test
- Purpose: Validate system reliability and sensitivity using human blood
- Protocol:
- Prepare 1% Intralipid solution suspended in 450 mL phosphate-buffered saline
- Add 5g yeast to solution for deoxygenation
- Introduce 0.5 mL human blood to solution
- Cycle between oxygenation (99.99% O₂ bubbling) and deoxygenation (yeast)
- Maintain temperature at 37°C throughout
- Monitor system response to hemoglobin concentration changes
Ink Drop Test
- Purpose: Evaluate system sensitivity to gradual concentration changes
- Protocol:
- Setup optical darkroom to eliminate ambient light
- Place probe in polyethylene container with 600 mL fresh water
- Maintain constant temperature at 31°C
- Inject 1 mL of 600-fold diluted carbon ink at 1-minute intervals
- Monitor system response to gradual absorption changes
- Ensure continuous stirring for homogeneous distribution
Multi-Channel Crosstalk Test
- Purpose: Quantify interference between adjacent channels
- Protocol:
- Activate single source channel while monitoring all detector channels
- Measure signal responses in non-target channels
- Calculate crosstalk ratio between target and non-target channels
- Repeat for all source-detector configurations
Hardware and Algorithmic Solutions
Crosstalk reduction requires both hardware design considerations and algorithmic approaches:
Hardware Design Strategies
- Increase trace spacing on PCB layouts to reduce electromagnetic coupling
- Use 45-degree fold lines instead of 90-degree angles in circuit board traces
- Implement short, thick traces for critical transmission lines
- Arrange adjacent layer traces perpendicular to minimize coupling
Algorithmic Solutions
- Employ subject-specific differential pathlength factor (DPF) estimation
- Utilize multi-distance high-density measurements to estimate DPF spectral dependence
- Apply advanced signal processing techniques to separate overlapping channel signals
Integrated EEG-fNIRS Processing Pipeline
For simultaneous EEG-fNIRS BCI research, an integrated processing pipeline effectively addresses artifacts in both modalities while leveraging their complementary nature.
Multimodal Data Fusion and Classification
The Multimodal DenseNet Fusion (MDNF) model represents an advanced approach for integrating EEG and fNIRS data [18]. This architecture effectively leverages the temporal richness of EEG and spatial specificity of fNIRS through several key steps:
EEG Processing Stream
- Channel Selection: Strategically select EEG channels based on neuroanatomical locations relevant to cognitive and motor functions
- Time-Frequency Transformation: Convert EEG signals into 2D time-frequency representations using Short-Time Fourier Transform
- Feature Extraction: Apply transfer learning with DenseNet to extract discriminative spatiotemporal features
fNIRS Processing Stream
- Signal Preprocessing: Convert raw intensity to optical density, then to hemoglobin concentrations using modified Beer-Lambert law
- Feature Extraction: Calculate spectral entropy features from fNIRS data
- Noise Removal: Apply appropriate filtering and artifact removal techniques
Multimodal Fusion
- Feature Integration: Combine EEG-derived image features with fNIRS spectral entropy features
- Classification: Implement customized DenseNet architecture for final task classification
- Validation: Achieve high classification accuracy across various cognitive and motor imagery tasks
This approach has demonstrated superiority over unimodal methods and other state-of-the-art fusion techniques in BCI applications [18].
Comprehensive Experimental Protocol for Simultaneous EEG-fNIRS
Equipment Setup
- EEG System Configuration:
- Position electrodes according to international 10-20 system
- Ensure impedance values below 5 kΩ for all electrodes
- Set appropriate sampling rate (≥250 Hz)
fNIRS System Configuration:
- Arrange sources and detectors for optimal coverage of regions of interest
- Include short-separation channels (≤1.5 cm) for noise reference
- Set appropriate sampling rate (≥10 Hz)
Synchronization:
- Implement hardware synchronization between EEG and fNIRS systems
- Record common trigger signals for both modalities
- Verify temporal alignment during preprocessing
Data Acquisition Protocol
- Baseline Recording: Collect 5-minute resting-state data for system calibration
- Task Paradigm: Implement event-related or block design based on research question
- Quality Monitoring: Continuously monitor signal quality during acquisition
- Artifact Annotation: Record potential artifact events (movements, exercises)
Integrated Processing Workflow
- Preprocessing:
- Apply modality-specific artifact removal techniques
- Perform temporal alignment and downsampling if necessary
- Extract epochs around events of interest
Feature Extraction:
- Extract temporal, spectral, and spatial features from EEG
- Calculate hemoglobin concentration changes and spatial patterns from fNIRS
- Generate complementary feature sets
Data Fusion:
- Implement appropriate fusion strategy (feature-level, decision-level)
- Apply machine learning or deep learning classification
- Validate using cross-validation and appropriate metrics
Integrated EEG-fNIRS Processing Pipeline
The Scientist's Toolkit: Research Reagents and Materials
Table 3: Essential Research Materials for Artifact Mitigation Studies
| Material/Reagent | Application | Function | Specifications |
|---|---|---|---|
| Intralipid Solution | fNIRS System Validation | Simulating tissue scattering properties | 1% suspension in phosphate-buffered saline |
| Human Blood Samples | Sensitivity Testing | Providing true tissue spectra and oxygenation capabilities | Fresh samples with anticoagulant |
| Carbon Ink | Sensitivity Testing | Providing monotonic absorption spectrum | Pure black ink, 600-fold dilution |
| Yeast | Deoxygenation Agent | Enzymatic oxygen consumption in blood models | 5g per 450mL solution |
| Phosphate-Buffered Saline | Solution Preparation | Maintaining physiological pH and osmolarity | Standard formulation, pH 7.4 |
| Optical Phantoms | System Calibration | Mimicking tissue optical properties | Polyethylene containers with defined absorption/scattering |
Effective mitigation of motion artifacts, physiological noise, and signal crosstalk is essential for maximizing the potential of simultaneous EEG-fNIRS systems in BCI research. The protocols and methodologies presented in this application note provide comprehensive approaches for addressing these challenges at various stages, from experimental design to data processing. By implementing these strategies, researchers can significantly improve signal quality and reliability, thereby enhancing the validity of their findings in both clinical and non-clinical applications. The continued development of advanced artifact handling techniques, particularly in the domain of multimodal data fusion and machine learning, promises to further expand the capabilities of simultaneous EEG-fNIRS systems in brain-computer interface research.
Advanced Channel Selection Techniques to Reduce Computational Burden
In simultaneous EEG-fNIRS brain-computer interface (BCI) research, channel selection has emerged as a critical preprocessing step for enhancing system performance. Channel selection directly influences computational efficiency, classification accuracy, and practical usability of hybrid BCI systems [50] [51]. The fundamental challenge stems from the high-dimensional data acquired through multiple EEG electrodes and fNIRS optodes, which creates computational bottlenecks without significantly improving discriminatory power [50] [52]. This application note examines advanced channel selection techniques that effectively reduce computational burden while maintaining—and in some cases enhancing—classification performance for motor imagery and cognitive tasks.
The complementary nature of EEG and fNIRS modalities creates unique opportunities for optimized channel selection. EEG provides millisecond-level temporal resolution for capturing rapid neural electrical activity, while fNIRS offers superior spatial localization of hemodynamic responses [53] [18]. However, this multimodal advantage comes with increased system complexity, as the combined channel sets can exceed 100 individual data sources [51]. Strategic channel selection addresses this limitation by identifying the most informative subsets of channels, thereby reducing redundant information and noise while preserving task-relevant neural signatures [50] [52].
Advanced Channel Selection Techniques
Correlation-Based Selection
The Pearson product-moment correlation coefficient (PPMCC) represents a statistically-grounded approach for hybrid EEG-fNIRS channel selection. This method quantifies linear associations between channels, ranking them according to their representation of true motor imagery signals versus noise or artifacts [50].
Key Protocol Steps:
- Data Preparation: Separate EEG and fNIRS channels into left and right hemisphere groups [50]
- Correlation Analysis: Calculate pairwise correlation coefficients (ρ) between channels using the formula: ρi,j = cov(i,j)/(σi × σ_j) [50]
- Rank Matrix Development: Create a rank matrix based on correlation strengths, where highest-ranked channels represent true motor imagery signals [50]
- Channel Selection: Select the most highly correlated channels from each hemisphere for further processing [50]
Performance Characteristics: Applied to a 21-channel EEG and 34-channel fNIRS setup, this approach significantly reduced computational burden while achieving classification accuracy comparable to full-channel sets when combined with KNN and Tree classifiers [50].
Meta-Heuristic and Evolutionary Algorithms
Evolutionary algorithms represent a sophisticated approach for identifying optimal channel subsets by simulating natural selection processes. These methods are particularly valuable for their ability to handle the non-linear, multi-objective optimization nature of channel selection [52] [54].
Table 1: Evolutionary Algorithms for Channel Selection
| Algorithm | Type | Key Features | Reported Performance |
|---|---|---|---|
| SPEA-II [54] | Multi-objective | Pareto front solutions, elitism preservation, nearest neighbor density estimation | Improved accuracy with 4.66 mean channels (similar to 8-channel set) |
| Dual-Front Sorting Algorithm (DFGA) [52] | Multi-objective discrete | Customized for BCI framework, computes solution sets with different channel counts | 3.9% accuracy improvement over common 8-channel P300 speller |
| Genetic Algorithm (GA) [52] | Single-objective | Population-based search, selection, crossover, mutation | Outperformed full channel sets in P300-based BCIs |
| Binary PSO (BPSO) [52] | Single-objective | Swarm intelligence, binary position representation | Effective for channel selection in P300 paradigms |
Implementation Workflow:
- Initialization: Create initial population of candidate channel subsets
- Fitness Evaluation: Assess each subset using objective functions (accuracy, channel count)
- Selection: Prioritize solutions dominating others in multi-objective space [54]
- Evolution Operations: Apply crossover and mutation to generate new solutions
- Elitism Preservation: Retain best-performing solutions across generations [54]
- Termination: Continue until convergence criteria met (e.g., stable Pareto front) [54]
Regularized Common Spatial Patterns with Optimization
The integration of Regularized Common Spatial Patterns (RCSP) with multi-objective optimization creates a powerful channel selection framework, particularly for motor imagery tasks. This approach combines the discriminative capability of CSP with the efficiency of evolutionary algorithms [54].
Key Advantages:
- Subject-Specific Selection: Tailors channel sets to individual users [54]
- Overfitting Prevention: Reduces redundant EEG channels and signal noise [54]
- Comfort Enhancement: Minimizes setup time and improves usability [54]
Experimental Results: Studies utilizing SPEA-II with RCSP demonstrated that typically 10-30% of total channels provided performance comparable to full-channel setups, dramatically reducing computational requirements while maintaining classification accuracy [51].
Experimental Protocols
Correlation-Based Channel Selection Protocol
Table 2: Protocol for PPMCC Channel Selection
| Step | Description | Parameters | Output |
|---|---|---|---|
| Data Acquisition | Simultaneous EEG-fNIRS recording during motor imagery tasks | EEG: 250Hz, 1-25Hz bandpass filter; fNIRS: 10.42Hz, 0.01-0.2Hz bandpass filter [50] | Raw EEG and fNIRS signals |
| Signal Preprocessing | Normalization and artifact removal | Subtract mean, divide by standard deviation [50] | Clean, normalized signals |
| Hemisphere Separation | Divide channels into left/right groups | Based on neuroanatomical positions [50] | Grouped channels for correlation analysis |
| Correlation Calculation | Compute PPMCC between channel pairs | ρ value range: [-1, 1] [50] | Correlation matrix |
| Channel Ranking | Create rank matrix based on correlation strength | Highest ρ = most representative [50] | Ranked channel list |
| Subset Selection | Select top-ranked channels from each hemisphere | Typically 10-30% of total channels [51] | Optimized channel subset |
Evolutionary Channel Selection Protocol
Table 3: Protocol for Meta-Heuristic Channel Selection
| Step | Description | Parameters | Output |
|---|---|---|---|
| Problem Formulation | Define optimization objectives | Minimize channel count, maximize accuracy [52] | Multi-objective problem framework |
| Algorithm Selection | Choose appropriate meta-heuristic | SPEA-II, DFGA, NSGA-II based on requirements [52] [54] | Selected algorithm with parameters |
| Population Initialization | Generate initial candidate solutions | Binary representation, random channel subsets [52] | Initial population |
| Fitness Evaluation | Assess each candidate subset | Classification accuracy, number of channels [52] | Fitness scores for all candidates |
| Evolutionary Operations | Apply selection, crossover, mutation | Tournament selection, uniform crossover [54] | New generation population |
| Termination Check | Evaluate stopping criteria | Fixed generations or convergence stability [52] | Final Pareto-optimal solutions |
| Solution Selection | Choose implementation-ready subset | Balance accuracy and practicality [52] | Optimal channel set for deployment |
Visualization of Workflows
Correlation-Based Channel Selection Workflow
Evolutionary Channel Selection Workflow
The Scientist's Toolkit
Table 4: Essential Research Reagents and Materials
| Item | Specification | Function/Purpose |
|---|---|---|
| EEG Amplifier | g.Nautilus, g.USBamp, or g.HIamp with 250+ Hz sampling [53] | Records electrical brain activity with high temporal resolution |
| fNIRS Sensor | g.SENSOR fNIRS or NIRSport2 with 760nm & 850nm wavelengths [53] | Measures hemodynamic responses via light absorption |
| Electrode Cap | Hybrid EEG/fNIRS cap with dark material, active electrodes [53] | Maintains fixed sensor position, prevents light leakage |
| Signal Processing Library | MATLAB, Python with MNE, BBCI Toolbox [50] [54] | Implements filtering, feature extraction, classification |
| Optimization Framework | Custom implementations of SPEA-II, NSGA-II, DFGA [52] [54] | Solves multi-objective channel selection problem |
| Validation Dataset | Public BCI repositories (BCI Competition, simultaneous EEG-fNIRS datasets) [50] [19] | Benchmarks algorithm performance across subjects |
Advanced channel selection techniques substantially enhance the practicality and performance of simultaneous EEG-fNIRS BCI systems. Correlation-based methods provide statistically robust approaches for identifying informative channels, while evolutionary algorithms offer powerful optimization frameworks for balancing competing objectives of accuracy and efficiency. The experimental protocols outlined in this application note provide researchers with practical methodologies for implementing these techniques, with typical implementations achieving 70-95% reduction in channel counts while maintaining classification accuracies of 80-95% across various motor imagery and cognitive tasks [50] [52] [51]. These approaches collectively address the critical computational challenges in hybrid BCI systems while paving the way for more practical, deployable brain-computer interface technologies.
Optimizing Feature Selection and Fusion Algorithms for Enhanced Performance
Electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS) are two non-invasive neuroimaging techniques that have gained significant traction in brain-computer interface (BCI) research. When used simultaneously, these modalities provide complementary information: EEG offers excellent temporal resolution for capturing fast neural electrical activity, while fNIRS provides superior spatial resolution for tracking slower hemodynamic responses [55]. However, effectively integrating these disparate signals requires sophisticated feature selection and fusion algorithms to overcome the inherent challenges of multimodal data processing and to enhance BCI performance for applications in neuroscience and clinical drug development.
This application note provides a comprehensive technical framework for optimizing feature selection and fusion methodologies in simultaneous EEG-fNIRS BCI systems. We present structured comparisons of algorithmic performance, detailed experimental protocols, and practical implementation tools to assist researchers in designing robust multimodal BCI studies for evaluating neurological function and pharmaceutical efficacy.
Performance Comparison of Feature Selection and Fusion Strategies
The selection of appropriate feature selection and fusion algorithms significantly impacts classification accuracy in EEG-fNIRS BCI systems. Based on comprehensive analysis of recent research, we have quantified the performance of various approaches to guide methodological decisions.
Table 1: Comparative Performance of Feature Selection Algorithms in EEG-fNIRS BCI
| Feature Selection Algorithm | Modality | Key Mechanism | Reported Accuracy | Reference |
|---|---|---|---|---|
| Binary Enhanced Whale Optimization Algorithm (E-WOA) | EEG-fNIRS | Wrapper-based approach with SVM cost function | 94.22% ± 5.39% | [56] |
| Genetic Algorithm (GA) | EEG-fNIRS | Non-linear feature selection with ensemble learning | 95.48% | [7] |
| Atomic Search Optimization | EEG-fNIRS | Multi-domain feature selection for progressive learning | 96.74% (MI), 98.42% (MA) | [55] |
| Particle Swarm Optimization (PSO) | EEG-only | Optimized channel selection for motor imagery | 76.7% | [57] |
Table 2: Performance of Fusion Strategies in Hybrid EEG-fNIRS BCI Systems
| Fusion Strategy | Fusion Level | Key Methodology | Advantages | Limitations |
|---|---|---|---|---|
| Early-Stage Fusion [58] | Data-level | Y-shaped neural network with shared layers | Preserves raw signal relationships; highest performance in comparative studies | High computational load; requires temporal alignment |
| Feature-level Fusion [56] [55] | Feature-level | Concatenation + optimized selection (E-WOA, ASO) | Balances information preservation & dimensionality reduction | Risk of feature redundancy without proper selection |
| Decision-level Fusion [55] [17] | Decision-level | Dempster-Shafer theory, classifier probability averaging | Robust to modality-specific noise; flexible implementation | Potentially lower accuracy than feature-level fusion |
| Deep Learning Fusion [17] | Hybrid | End-to-end learning with attention mechanisms | Automatic feature extraction; minimal manual engineering | Requires large datasets; limited interpretability |
Experimental Protocols for EEG-fNIRS Feature Optimization
Protocol: Optimization-Based Feature Selection with E-WOA
This protocol implements a wrapper-based feature selection approach for discriminating motor imagery tasks using hybrid EEG-fNIRS features [56].
Materials and Setup:
- Simultaneous EEG-fNIRS recording system
- 30 EEG electrodes (10-5 international system) and 36 fNIRS channels (14 sources, 16 detectors)
- Processing environment: MATLAB or Python with optimization toolboxes
Procedure:
- Data Acquisition:
- Record EEG at 1000 Hz and fNIRS at 2.5 Hz during motor imagery tasks (e.g., left-hand vs. right-hand imagery)
- Structure experiment with 60s initial rest, 20 trials per session, with 10s task periods and 15-17s random rest intervals
Preprocessing:
- For EEG: Apply common average referencing, bandpass filter (0.5-50 Hz), and remove EOG artifacts using hybrid ICA-regression [56]
- For fNIRS: Apply third-order Butterworth bandpass filter (0.01-0.1 Hz) to extract hemodynamic components
Feature Extraction:
- Calculate temporal statistical features (mean, variance, slope) for both modalities using 10s sliding windows
- For EEG: Extract band power features (μ-band: 8-13 Hz, β-band: 13-30 Hz) from motor cortex channels
- For fNIRS: Compute ΔHbO and ΔHbR concentration changes from all channels
Feature Fusion and Selection:
- Concatenate all EEG and fNIRS features into a unified feature vector
- Initialize E-WOA with SVM-based cost function for fitness evaluation
- Set population size to 50 and maximum iterations to 100
- Execute binary E-WOA to select optimal feature subset maximizing classification accuracy
Validation:
- Implement k-fold cross-validation (k=10) to assess generalizability
- Compare performance against conventional WOA and other optimization algorithms
Protocol: Multi-Domain Feature Fusion with Progressive Learning
This protocol implements a comprehensive fusion approach combining multi-domain features with progressive learning for enhanced classification of motor imagery and mental arithmetic tasks [55].
Materials and Setup:
- EEG system with minimum 200 Hz sampling rate
- fNIRS system with minimum 10 Hz sampling rate
- Processing environment supporting deep learning frameworks (TensorFlow/PyTorch)
Procedure:
- Multi-Domain Feature Extraction:
- EEG Features:
- Temporal: Statistical moments (mean, variance, skewness, kurtosis)
- Spectral: Band power (δ, θ, α, β, γ), spectral edge frequency
- Time-Frequency: Wavelet coefficients, Hilbert-Huang transform
- fNIRS Features:
- Temporal: ΔHbO/ΔHbR mean, slope, peak, area under curve
- Spatial: Channel-wise topographic features
- Morphological: Signal rise time, fall time, hysteresis
- EEG Features:
Feature Selection:
- Implement Atomic Search Optimization for feature subspace selection
- Configure ASO population size to 30 with 50 iterations
- Use maximum relevance minimum redundancy criterion for fitness function
Progressive Fusion Architecture:
- Level 1: Modality-specific feature learning with dedicated autoencoders
- Level 2: Cross-modal correlation analysis with canonical correlation analysis
- Level 3: Joint representation learning through fully connected layers
Classification and Validation:
- Implement ensemble classifier (SVM, Random Forest, KNN) with stacking
- Validate using leave-one-subject-out cross-validation
- Compare against single-modality and single-domain baselines
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Essential Research Materials for EEG-fNIRS BCI Studies
| Item | Specifications | Research Function | Example Applications |
|---|---|---|---|
| EEG Recording System | 30+ channels, 1000+ Hz sampling, Ag/AgCl electrodes | Records electrophysiological brain activity with high temporal resolution | Motor imagery detection, ERD/ERS analysis [56] |
| fNIRS Recording System | 30+ channels, 2.5+ Hz sampling, 760nm & 850nm wavelengths | Monitors hemodynamic responses via HbO/HbR concentration changes | Localizing cortical activation, complementing EEG [59] |
| Simultaneous Recording Cap | Integrated EEG electrodes & fNIRS optodes, 10-5 system placement | Ensures precise colocation of multimodal sensors for correlated data | Multimodal data acquisition with spatial correspondence [60] |
| Artifact Removal Tools | ICA algorithms, regression methods, motion correction | Removes physiological and motion artifacts from neural signals | Improving signal quality for feature extraction [56] |
| Optimization Toolboxes | MATLAB Optimization, PyGMO, DEAP, custom algorithms | Implements feature selection algorithms (WOA, GA, PSO, ASO) | Dimensionality reduction, optimal feature subset selection [56] [55] |
| Deep Learning Frameworks | TensorFlow, PyTorch, custom neural network architectures | Implements fusion networks and end-to-end learning | Advanced feature fusion, classification [55] [58] |
The strategic implementation of feature selection and fusion algorithms is paramount for maximizing the performance of simultaneous EEG-fNIRS BCI systems. Optimization-based feature selection approaches, particularly the Enhanced Whale Optimization Algorithm and Atomic Search Optimization, demonstrate significant advantages over conventional methods, achieving classification accuracies exceeding 94% for motor imagery tasks. Furthermore, the fusion strategy selection critically impacts outcomes, with early-stage fusion providing potential performance benefits at the cost of computational complexity, while feature-level fusion with proper selection offers an effective balance for practical implementations.
These protocols and analyses provide researchers and drug development professionals with validated methodologies for implementing optimized EEG-fNIRS systems. The structured comparison of algorithmic performance enables informed selection of appropriate strategies for specific research objectives, particularly in clinical trials and neuropharmaceutical efficacy studies where robust biomarker detection is essential.
Hardware and Software Solutions for Improved Scalp Coupling and Signal Integrity
Functional near-infrared spectroscopy (fNIRS) and electroencephalography (EEG) are non-invasive neuroimaging techniques that, when combined, provide a powerful multimodal tool for brain-computer interface (BCI) research. fNIRS measures hemodynamic responses by detecting changes in oxyhemoglobin (HbO) and deoxyhemoglobin (HbR) concentrations, while EEG records the brain's electrical activity with high temporal resolution [1]. A significant challenge in acquiring high-quality data, especially for simultaneous EEG-fNIRS setups, is ensuring optimal scalp coupling—the quality of contact between optodes/electrodes and the scalp. Poor coupling introduces noise and artifacts, compromising signal integrity and the reliability of subsequent neural decoding [61] [62]. This application note details standardized protocols and solutions to overcome these challenges, framed within the context of a robust BCI research framework.
Quantitative Metrics for Signal Quality Assessment
Objective metrics are essential for quantifying signal quality during setup and acquisition. The following parameters should be monitored in real-time.
Table 1: Key Quantitative Metrics for fNIRS Signal Quality Assurance
| Metric | Definition | Calculation Method | Target Value | Interpretation |
|---|---|---|---|---|
| Scalp Coupling Index (SCI) [61] [63] | Quantifies prominence of cardiac signal in fNIRS raw data. | Correlation between cardiac pulsations (0.5-2.5 Hz) in two wavelength signals. | SCI ≥ 0.8 [63] | Values ≥ 0.9 are "good"; < 0.8 indicates poor coupling and channel should be rejected [63]. |
| Signal-to-Noise Ratio (SNR) [61] | Objective measure of signal quality relative to noise. | Combines SCI with additional power features of the photoplethysmographic signal. | Maximize | A higher SNR indicates a cleaner signal, sufficient for reliable cortical hemodynamic estimation [61]. |
| Gain/Amplitude [63] | Instrument amplifier setting to achieve detectable light intensity. | Set during calibration. | Within dynamic range, avoiding overexposure | Overexposure, indicated by a straight wave with spikes, requires optode repositioning or gain readjustment [63]. |
| Heart Rate Peak [63] | Visual identification of cardiac pulsation in raw signal. | Visual inspection of ~1 Hz oscillations in raw light intensity or optical density. | Clearly visible | Confirms adequate signal penetration and coupling. Absence suggests poor contact or obstruction [63]. |
Experimental Protocols for Optimal Setup and Data Acquisition
Protocol: Pre-Acquisition Headgear Preparation and Optode Coupling
This protocol aims to minimize setup time and maximize the number of usable channels by ensuring optimal optode-scalp contact [61] [63].
Research Reagents & Materials:
- fNIRS System: A continuous-wave (CW) fNIRS system (e.g., NIRSport, NIRScout) compatible with real-time monitoring software [61].
- EEG System: A compatible EEG system (e.g., 128-channel Electrical Geodesics system) integrated within the fNIRS cap [64].
- Optodes: Light source and detector optodes appropriate for the system.
- Coupling Gel: (For wet systems) Electrolyte gel for EEG electrodes; fNIRS may use gels or direct contact.
- Hair Parting Tools: Blunt-ended plastic tools, syringe plungers, or specialized combs for parting hair [63].
- Real-Time Monitoring Software: Software such as PHOEBE (Placing Headgear Optodes Efficiently Before Experimentation) or manufacturer-specific tools [61].
Methodology:
- Headgear Sizing and Placement: Select an appropriately sized EEG/fNIRS cap based on the participant's head circumference. Position the cap according to the international 10-20 system, aligning optodes over the regions of interest (e.g., sensorimotor and parietal cortices for AON studies) [64].
- Hair Management: Part the hair meticulously underneath each optode location to ensure direct skin contact. For individuals with thick, dark, or curly hair, this step is critical as hair blocks light and increases absorption [63].
- Optode Adjustment: Seat each optode firmly against the scalp. Using real-time monitoring software (e.g., PHOEBE), observe the coupling status of all individual optodes displayed on a head model.
- Real-Time Quality Control: Initiate a short data acquisition. The software will compute the SCI/SNR for each channel in real time. Optodes connected to channels with low SCI will be visually highlighted, guiding the experimenter to make targeted adjustments [61].
- Iterative Optimization: Adjust the highlighted optodes (e.g., reposition, part hair further) and observe the real-time feedback until the SCI for all channels meets the target threshold (≥ 0.8). This process significantly shortens the pre-acquisition preparation time.
Protocol: Post-Acquisition fNIRS Data Preprocessing for BCI
This protocol outlines a standard pipeline for converting raw fNIRS data into clean hemoglobin concentration changes suitable for BCI feature extraction, based on established practices in tools like MNE-Python [42].
Research Reagents & Materials:
- Computing Environment: A computer with data processing software (e.g., MNE-Python, Homer2, or custom scripts in MATLAB/Python).
- Processed Data: Raw intensity data acquired from the fNIRS system.
Methodology:
- Channel Exclusion: Reject channels with poor scalp coupling based on the pre-acquisition SCI assessment or visual inspection of the raw signal [42] [61].
- Intensity to Optical Density: Convert the raw light intensity data to optical density (OD) to linearize the signal and reduce instrumental dependencies.
raw_od = mne.preprocessing.nirs.optical_density(raw_intensity)[42]. - Optical Density to Hemoglobin: Apply the Modified Beer-Lambert Law (MBLL) to convert OD changes to relative changes in HbO and HbR concentration. A typical partial pathlength factor (PPF) is 0.1.
raw_haemo = mne.preprocessing.nirs.beer_lambert_law(raw_od, ppf=0.1)[42]. - Filtering: Apply a band-pass filter (e.g., 0.05 - 0.7 Hz) to the hemoglobin data. This removes high-frequency noise (e.g., cardiac pulsation ~1 Hz) and very low-frequency drift, isolating the hemodynamic response of interest for most BCI tasks [42].
Diagram 1: fNIRS data preprocessing workflow for BCI applications.
Protocol: Simultaneous EEG-fNIRS Data Fusion and Analysis
This protocol describes a method for integrating the preprocessed EEG and fNIRS data to leverage their complementary information, as demonstrated in studies of the Action Observation Network (AON) [64].
Research Reagents & Materials:
- Multimodal Dataset: Time-synchronized and preprocessed EEG and fNIRS data.
- Analysis Software: Advanced computational tools capable of multimodal fusion (e.g., structured sparse multiset Canonical Correlation Analysis - ssmCCA - toolboxes in MATLAB or Python) [64].
Methodology:
- Unimodal Analysis: Analyze EEG and fNIRS data separately to identify condition-related neural activity (e.g., Event-Related Desynchronization in EEG, HbO activation in fNIRS). This provides a baseline for evaluating the fused results [64].
- Data Fusion with ssmCCA: Apply ssmCCA to the unimodal datasets. This method hierarchically extracts deep features from both modalities and finds a shared representation by identifying canonical variables that maximize the correlation between the two datasets [64].
- Interpretation: The fused output pinpoints brain regions where both electrical (EEG) and hemodynamic (fNIRS) activities are consistently correlated. This provides a more robust and validated map of neural engagement than unimodal analyses alone, which may show discrepancies [64].
Diagram 2: Simultaneous EEG-fNIRS data fusion and analysis process.
The Scientist's Toolkit
Table 2: Essential Hardware and Software for EEG-fNIRS BCI Research
| Tool Name | Type | Primary Function | Key Feature for BCI |
|---|---|---|---|
| NIRSport2 [65] | fNIRS Acquisition Hardware | Measures cortical hemodynamics (HbO/HbR). | Portability, compatibility with EEG, integrated with Lab Streaming Layer (LSL) for real-time BCI/Neurofeedback [65]. |
| Aurora fNIRS [65] | fNIRS Acquisition Software | Controls NIRx fNIRS instruments and acquires data. | Automated signal optimization and real-time visualization of HbO/HbR, suited for neurofeedback paradigms [65]. |
| PHOEBE [61] | Signal Quality Software | Computes SCI/SNR and visualizes optode coupling in real-time. | Drastically reduces setup time and maximizes usable channels by identifying poorly coupled optodes before data acquisition [61]. |
| MNE-Python [42] | Data Analysis Software | Open-source Python package for processing M/EEG and fNIRS data. | Provides a complete, standardized pipeline from raw data to evoked responses and source modeling, ensuring reproducible preprocessing [42]. |
| Turbo-Satori [65] | Real-Time Analysis Software | Real-time fNIRS analysis for BCI and Neurofeedback. | User-friendly interface for designing real-time fNIRS experiments and processing streams, enabling immediate feedback [65]. |
| Structured Sparse Multiset CCA (ssmCCA) [64] | Advanced Analysis Algorithm | Fuses multimodal EEG and fNIRS data. | Identifies brain regions with consistent activity across modalities, enhancing the validity and spatial specificity of BCI control signals [64]. |
Achieving high-fidelity data in simultaneous EEG-fNIRS experiments is contingent upon rigorous attention to scalp coupling and signal integrity. The protocols and tools detailed herein provide a concrete framework for researchers to reliably set up their systems, preprocess data, and fuse multimodal signals. By systematically implementing these hardware and software solutions, BCI research can advance with greater methodological rigor, leading to more robust and interpretable findings on brain function and improved real-time applications.
Validating Performance: Benchmarking Hybrid BCI Against Unimodal Systems
In Brain-Computer Interface (BCI) research, the integration of electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS) has emerged as a powerful multimodal approach to overcome the limitations of unimodal systems [66] [1]. This integration capitalizes on the complementary strengths of EEG's millisecond-scale temporal resolution and fNIRS's superior spatial specificity for hemodynamic responses [64] [18]. Quantitative performance metrics are essential for evaluating the efficacy of these hybrid systems in clinical and research applications, particularly for motor imagery (MI) tasks relevant to neurorehabilitation and drug development research [66] [67]. This document outlines the critical metrics, provides experimental protocols, and offers a research toolkit for implementing simultaneous EEG-fNIRS BCIs.
Performance Metrics for EEG-fNIRS BCIs
The table below summarizes key quantitative metrics reported in recent EEG-fNIRS BCI studies, highlighting the performance advantages of multimodal fusion approaches over unimodal systems.
Table 1: Performance Metrics in EEG-fNIRS BCI Studies
| Study Reference | Modality & Approach | Classification Accuracy (%) | Information Transfer Rate (bits/min) | Key Application Context |
|---|---|---|---|---|
| PMC12631896 (2025) | Hybrid EEG-fNIRS with Transfer Learning | 74.87 (patient data); 82.30 & 87.24 (public datasets) | Not Reported | Intracerebral Hemorrhage (ICH) Rehabilitation [66] |
| IEEE JTEHM (2024) | Multimodal DenseNet Fusion (MDNF) | Superior to referenced state-of-the-art methods | Not Reported | Motor Imagery & Cognitive Tasks [18] |
| NeuroImage (2012) | Hybrid NIRS-EEG Meta-classifier | +5% average improvement | Hindered by hemodynamic delay | Sensory Motor Rhythm (SMR) BCI [38] |
| Sci Rep (2023) | Structured Sparse Multiset CCA | Differentiated activation between conditions | Not Reported | Motor Execution, Observation, and Imagery [64] |
| Frontiers Hum Neurosci (2025) | Heterogeneous Transfer Learning (CHTLM) | 83.1 (pre-rehab); 91.3 (post-rehab) | Not Reported | Cross-subject MI classification in stroke [67] |
| BOE (2016) | jICA-based EEG-fNIRS fusion | Improved +3.4% vs EEG; +11% vs fNIRS | Not Reported | Mental Stress Assessment [68] |
Classification Accuracy remains the most widely reported metric, demonstrating consistent improvements in hybrid systems compared to unimodal approaches [66] [38]. The Information Transfer Rate (ITR), which quantifies the speed and accuracy of communication, is less frequently reported but remains crucial for real-time BCI applications [69]. Factors influencing these metrics include the fusion strategy (early, late, hybrid), feature extraction methods, and classification algorithms [66] [18].
Experimental Protocols for EEG-fNIRS Motor Imagery Paradigms
Protocol 1: Motor Imagery for Stroke Rehabilitation
This protocol is adapted from studies involving patients with intracerebral hemorrhage or stroke [66] [67].
Objective: To acquire synchronized EEG-fNIRS data during motor imagery tasks for developing rehabilitation BCIs. Participants: Normal controls and patients with motor impairments (e.g., post-stroke). Typical cohort sizes range from 13-30 participants [66] [64]. Equipment:
- Simultaneous EEG-fNIRS recording system
- EEG cap integrated with fNIRS optodes (e.g., 64-channel EEG + 48-channel fNIRS) [70]
- Display screen for task instructions
- Comfortable chair in a controlled environment
Procedure:
- Preparation: Position participant 60-80 cm from the screen. Apply conductive gel for EEG and ensure proper fNIRS optode-scalp contact.
- Baseline Recording: Record a 5-minute resting-state baseline with eyes open.
- Task Paradigm (Block Design):
- Cue Phase (2 s): A visual or auditory cue indicates the upcoming task.
- Imagery Phase (6-15 s): Participant performs kinesthetic motor imagery (e.g., imagining grasping a cup with the affected hand without actual movement). The duration can vary by protocol [66] [67].
- Rest Phase (10-20 s): Participant relaxes; cross-fixation point displayed.
- Trial Repetition: Repeat steps 3 for 20-48 trials per session to ensure adequate data [67].
- Data Synchronization: Use hardware triggers to synchronize EEG, fNIRS, and task event markers.
Data Analysis:
- EEG Processing: Bandpass filter (e.g., 0.5-40 Hz), artifact removal (e.g., ICA), and extract event-related desynchronization/synchronization (ERD/ERS) in mu/beta rhythms [66].
- fNIRS Processing: Convert raw light intensity to oxygenated (HbO) and deoxygenated hemoglobin (HbR) concentrations using the modified Beer-Lambert law. Bandpass filter (0.01-0.2 Hz) to remove physiological noise [1] [67].
- Feature Fusion & Classification: Implement transfer learning or deep learning models (e.g., CNNs, DenseNet) to fuse EEG and fNIRS features for classification [66] [18] [67].
Figure 1: Motor Imagery Task Workflow. This block design shows the sequence of phases in a single trial, repeated multiple times during an experiment.
Protocol 2: Multimodal Fusion for Cognitive State Classification
Objective: To classify cognitive states (e.g., mental stress, workload) using hybrid EEG-fNIRS features for applications in neuroergonomics and drug development [68].
Procedure:
- Task Design: Implement a mental arithmetic task with time pressure and negative feedback to induce stress [68].
- Paradigm: Use a block design with alternating task (e.g., 30 s of arithmetic) and rest (20 s) periods.
- Data Acquisition: Simultaneously record EEG (focusing on frontal theta and alpha power) and fNIRS (from prefrontal cortex HbO).
- Feature Fusion: Apply joint Independent Component Analysis (jICA) or canonical correlation analysis (CCA) to fuse EEG spectral power and fNIRS HbO features [68] [18].
- Classification: Train a support vector machine (SVM) or deep neural network (DNN) to discriminate between high and low cognitive load states.
The Scientist's Toolkit: Research Reagents & Materials
Table 2: Essential Materials for Simultaneous EEG-fNIRS Research
| Item | Specification/Function | Research Context |
|---|---|---|
| EEG System | High-density (≥64 channels) amplifier with active electrodes; reduces environmental noise. | Electrical neural activity recording with millisecond resolution [18] [70]. |
| fNIRS System | Continuous-wave system with dual wavelengths (730 & 850 nm); measures HbO and HbR concentration changes. | Hemodynamic activity monitoring with centimeter-scale spatial resolution [1] [67]. |
| Integrated Cap | EEG electrodes embedded with fNIRS optodes in a single cap; ensures co-registration of measurement locations. | Enables precise spatial correlation of electrical and hemodynamic responses [64] [70]. |
| Data Sync Unit | Hardware trigger box or software interface; generates simultaneous event markers for both systems. | Critical for temporal alignment of EEG and fNIRS data streams [1]. |
| Conductive Gel | Electrolyte gel for EEG; lowers electrode-scalp impedance for high-quality signal acquisition. | Essential for obtaining low-noise EEG data [70]. |
| 3D Digitizer | Magnetic or optical system (e.g., Polhemus Fastrak); records precise 3D optode/electrode coordinates. | Allows for co-registration with anatomical brain images [64]. |
Signaling Pathways and Computational Workflow
The analytical process for deriving quantitative metrics from raw EEG-fNIRS data involves a multi-stage computational pipeline, as illustrated below.
Figure 2: EEG-fNIRS Data Analysis Pipeline. The workflow from raw data to performance metrics, highlighting parallel processing streams for each modality that converge at the fusion stage.
Key Signaling Pathways and Neural Correlates:
- Action Observation Network (AON): Activated during motor execution, observation, and imagery. Key nodes include the premotor cortex, inferior frontal gyrus, and inferior parietal lobule, identifiable via fused EEG-fNIRS activation patterns [64] [70].
- Neurovascular Coupling: The biological link between electrical activity (EEG) and hemodynamic response (fNIRS). Simultaneous measurement allows for investigating this relationship, improving neural signal estimation [1].
- Prefrontal Cortex (PFC) Activation: fNIRS-measured HbO increases in the PFC during cognitive tasks like mental arithmetic, while EEG shows decreased frontal alpha power, indicating increased cognitive load or stress [68].
The quantitative evaluation of simultaneous EEG-fNIRS systems demonstrates clear advantages over unimodal BCIs, primarily through enhanced classification accuracy for motor imagery and cognitive tasks. The consistent reporting of accuracy metrics across studies underscores its reliability, while ITR requires more systematic assessment for real-time applications. The experimental protocols and research toolkit provided here offer a foundation for standardized implementation in clinical and pharmaceutical research settings, particularly for rehabilitation engineering and cognitive state monitoring. Future work should focus on standardizing ITR reporting and developing more efficient real-time fusion algorithms to further improve BCI performance and reliability.
Brain-Computer Interface (BCI) technology has evolved significantly from unimodal systems to sophisticated multimodal architectures. This application note provides a comparative analysis of hybrid electroencephalography (EEG)-functional near-infrared spectroscopy (fNIRS) systems against traditional EEG-only or fNIRS-only implementations. Through structured performance evaluation tables, detailed experimental protocols, and technical implementation guidelines, we demonstrate how the synergistic integration of electrophysiological and hemodynamic signals enhances classification accuracy, robustness, and applicability across diverse BCI paradigms. The complementary temporal and spatial profiles of EEG and fNIRS enable hybrid systems to overcome fundamental limitations inherent in unimodal approaches, offering researchers a validated framework for advancing brain-computer interface capabilities in both clinical and experimental settings.
EEG and fNIRS measure distinct yet complementary aspects of brain activity. EEG records electrical potentials generated by synchronized neuronal firing with excellent temporal resolution (milliseconds) but limited spatial resolution and susceptibility to physiological artifacts [71] [1]. Conversely, fNIRS measures hemodynamic responses through near-infrared light absorption by hemoglobin species, providing superior spatial localization but slower temporal response due to neurovascular coupling delays [1] [72]. This neurovascular coupling, where neuronal activity triggers localized blood flow changes, forms the physiological basis for fNIRS signal generation [72].
Hybrid EEG-fNIRS systems strategically leverage these complementary properties to create BCIs with enhanced capabilities. The electrical activity captured by EEG provides immediate detection of neural events, while the hemodynamic response measured by fNIRS offers improved spatial specificity and resilience to artifacts that often contaminate EEG signals [2] [1]. This multimodal approach enables more accurate classification of brain states by providing orthogonal information streams from the same neural substrates, allowing for cross-validation and richer feature extraction for machine learning algorithms [13].
The integration of these modalities is particularly valuable for studying complex cognitive-motor processes involving distributed networks like the Action Observation Network (AON), where combined electrical and hemodynamic monitoring provides more complete characterization of neural dynamics during motor execution, observation, and imagery tasks [64].
Performance Comparison: Quantitative Analysis
Table 1: Classification Accuracy Comparison Across Modalities
| Task Type | EEG-only | fNIRS-only | Hybrid EEG-fNIRS | Improvement vs. Unimodal | Citation |
|---|---|---|---|---|---|
| Motor Execution (Left vs. Right Hand) | 85.64% ± 7.4% | 85.55% ± 10.72% | 91.02% ± 4.08% | +5.38% (EEG), +5.47% (fNIRS) | [71] |
| Motor Imagery (General) | ~65% (Baseline) | ~65% (Baseline) | 70.18% (Average) | +5.18% | [73] |
| Mental Arithmetic | ~81% (Baseline) | ~81% (Baseline) | 86.26% (Average) | +5.26% | [73] |
| Word Generation | ~76% (Baseline) | ~76% (Baseline) | 81.13% (Average) | +5.13% | [73] |
| Force/Speed Motor Imagery | ~84% (EEG-only est.) | ~84% (fNIRS-only est.) | 89% ± 2% | +5% | [74] |
| Multiple Motor Tasks (4-class) | Limited in unimodal | Limited in unimodal | Significantly enhanced | Enables complex classification | [60] |
Table 2: Technical Characteristics Comparison
| Parameter | EEG-only | fNIRS-only | Hybrid EEG-fNIRS |
|---|---|---|---|
| Temporal Resolution | Excellent (ms) | Limited (1-2s) | Excellent (via EEG) |
| Spatial Resolution | Limited (2-3cm) | Good (1-2cm) | Good (via fNIRS) |
| Signal Origin | Electrical neuronal activity | Hemodynamic response | Both electrical & metabolic |
| Artifact Sensitivity | High (EMG, EOG, line noise) | Low (robust to electrical artifacts) | Complementary robustness |
| Setup Complexity | Moderate | Moderate | High (integrated systems) |
| Portability | High | High | High |
| Information Transfer Rate | Moderate | Slow | Enhanced |
| Source Localization | Challenging | Good | Improved through fusion |
| Delayed Response Analysis | Not applicable | Fundamental limitation | Compensated via EEG |
| Initial Cost | Low-moderate | Low-moderate | Moderate-high |
Experimental Protocols
Protocol 1: Motor Imagery Classification
Objective: To classify imagined hand movements for BCI control applications.
Experimental Setup:
- Participants: 15 healthy right-handed subjects [60]
- Task: Four-class motor execution/imagery (right-arm, left-arm, right-hand, left-hand)
- Trial Structure: 6s rest, 6s task execution, randomized across 25 trials per class
- Equipment: 21-channel EEG system (250Hz) + 34-channel fNIRS (10.42Hz) with wavelengths at 760nm and 850nm [60]
Signal Acquisition:
- EEG Parameters: Electrodes placed over motor cortex (F3, Fz, F4, FC5, FC1, FC2, FC6, C5, C3, C1, Cz, C2, C4, C6, CP5, CP1, CP2, CP6, P3, Pz, P4), referenced to FCz, ground at Fpz [60]
- fNIRS Parameters: 12 sources, 12 detectors arranged over motor cortex with maximum 3.4cm source-detector distance to ensure signal quality [60]
Signal Processing:
- EEG Processing: Bandpass filtering (0.5-40Hz), Common Spatial Patterns (CSP) for feature extraction
- fNIRS Processing: Conversion of optical density to hemoglobin concentrations using Modified Beer-Lambert Law, bandpass filtering (0.01-0.2Hz) to remove physiological noise [50]
- Feature Fusion: Regularized CSP optimized with genetic algorithms, combining EEG spatial patterns with fNIRS slope indicators to reduce hemodynamic response delay [60]
Classification: Linear Discriminant Analysis or Support Vector Machines for multi-class discrimination.
Protocol 2: Mental Task Classification
Objective: To discriminate between different cognitive states for neuroergonomics and clinical applications.
Experimental Setup:
- Participants: 29 healthy subjects (14 male, 15 female) [73]
- Tasks: Motor Imagery (MI), Mental Arithmetic (MA), and Word Generation (WG)
- Equipment: 30-channel EEG (1000Hz) + fNIRS system
Advanced Fusion Methodology:
- TSFNet Architecture: Implements temporal-spatial fusion through two specialized layers [73]
- EEG-fNIRS-guided Fusion (EFGF) Layer: Extracts temporal features from EEG and spatial features from fNIRS to generate hybrid attention maps
- Cross-Attention-based Feature Enhancement (CAFÉ) Layer: Enables bidirectional interaction between fNIRS and fusion features via cross-attention mechanism
- Implementation: Achieves accuracies of 70.18% (MI), 86.26% (MA), and 81.13% (WG) [73]
Protocol 3: Asynchronous BCI With Early Response Detection
Objective: To minimize system latency by detecting early features in both modalities.
Experimental Approach:
- EEG Features: Focus on 0-1s post-stimulus interval for rapid response detection [71]
- fNIRS Features: Extract initial dip (0-2s) instead of full hemodynamic response [71]
- Channel Selection: Employ Pearson correlation coefficient or General Linear Model (GLM) to identify most informative channels [71] [50]
- Advantage: Reduces command delay from typical 6-7.5s to 2-3s, enabling more natural BCI interaction
Technical Implementation Guidelines
System Integration Architecture
Fusion Methodologies
A. Data-Level Fusion:
- Structured sparse multiset Canonical Correlation Analysis (ssmCCA) to identify shared neural components [64]
- Applications: Identifying Action Observation Network activation across motor execution, observation, and imagery
B. Feature-Level Fusion:
- Temporal-phase-frequency features from EEG combined with HbD (HbO-HbR) features from fNIRS [74]
- Joint Mutual Information (JMI) criterion for feature selection and optimization
- Extreme Learning Machines (ELM) for classification achieving 89% accuracy [74]
C. Decision-Level Fusion:
- Weighted combination of classifier outputs from unimodal streams
- Adaptive weighting based on signal quality and task demands
Hybrid Cap Design and Co-registration
Integrated Assembly:
- Base EEG cap (e.g., actiCAP 128) with perforations for fNIRS optode mounting [60]
- 3D-printed customizable helmets for precise optode positioning [2]
- Cryogenic thermoplastic sheets for subject-specific customization [2]
Spatial Configuration:
- fNIRS source-detector distances: 2.5-3.5cm for adult studies [1] [60]
- EEG electrode placement following international 10-20 system
- Co-registration using 3D digitization (e.g., Fastrak) for anatomical localization [64]
The Scientist's Toolkit: Essential Research Reagents
Table 3: Essential Equipment and Analytical Tools
| Category | Item | Specification/Function | Representative Examples |
|---|---|---|---|
| Acquisition Hardware | EEG System | High-temporal resolution electrical signal acquisition | BrainAmp DC, microEEG |
| fNIRS System | Hemodynamic response measurement via NIR light | NIRScout, Hitachi ETG-4100 | |
| Integrated Caps | Simultaneous mounting of EEG electrodes and fNIRS optodes | actiCAP + custom modifications | |
| Analytical Tools | Preprocessing | Signal filtering, artifact removal | EEGLAB, NIRS-KIT |
| Feature Extraction | Temporal, spatial, frequency feature identification | CSP, AR models, Wavelet transforms | |
| Fusion Algorithms | Multimodal integration | ssmCCA, TSFNet, RCSP | |
| Classification | Pattern recognition and BCI control | SVM, LDA, ELM, Deep Learning | |
| Validation Metrics | Performance | Classification accuracy, Information Transfer Rate | Cross-validation protocols |
| Signal Quality | Signal-to-noise ratio, artifact quantification | Visual inspection, automated metrics |
Signaling Pathways and Experimental Workflow
Hybrid EEG-fNIRS systems represent a significant advancement over unimodal BCI approaches, consistently demonstrating superior classification accuracy across motor imagery, mental arithmetic, and other cognitive tasks. The complementary nature of electrophysiological and hemodynamic signals enables researchers to overcome fundamental limitations of either modality alone, particularly the trade-off between temporal and spatial resolution.
Future development directions include:
- Miniaturization of integrated systems for real-world applications
- Advanced deep learning architectures for automated feature learning and fusion
- Real-time adaptive algorithms that dynamically weight modality contributions based on signal quality
- Expanded clinical applications in neurological rehabilitation, disorder assessment, and brain state monitoring
The protocols and methodologies outlined in this application note provide researchers with comprehensive guidelines for implementing hybrid EEG-fNIRS systems, supported by empirical evidence of performance advantages over traditional unimodal approaches.
Simultaneous Electroencephalography (EEG) and functional Near-Infrared Spectroscopy (fNIRS) has emerged as a powerful multimodal neuroimaging technique with significant potential for both clinical diagnostics and brain-computer interface (BCI) development. This integration capitalizes on the complementary strengths of each modality: EEG provides millisecond-level temporal resolution of electrical brain activity, while fNIRS offers superior spatial localization of hemodynamic responses associated with neural activation [1] [75]. The portability, cost-effectiveness, and tolerance to motion artifacts of this combined system make it particularly suitable for studying dynamic brain states in clinical populations and for developing real-world BCI applications [1] [76].
Within the framework of BCI research, this multimodal approach addresses critical limitations of unimodal systems. EEG-based BCIs, while excellent for tracking rapid neural dynamics, suffer from limited spatial specificity and susceptibility to signal interference. Conversely, fNIRS-based BCIs provide valuable spatial information but are constrained by the inherent latency of the hemodynamic response [18]. Their integration creates a more robust feature space, enhancing classification accuracy across a wider range of tasks and user states [18] [75]. This technical note details the application of simultaneous EEG-fNIRS through specific clinical case studies and provides standardized protocols for its implementation in research on neurological and neuropsychiatric conditions.
Case Study Applications and Quantitative Findings
The application of simultaneous EEG-fNIRS across various clinical domains has yielded quantitative insights into brain function and pathophysiology. The table below summarizes key findings from research in relevant cognitive and neurological conditions.
Table 1: Quantitative Findings from EEG-fNIRS Studies in Relevant Domains
| Domain / Task | Experimental Paradigm | EEG Findings | fNIRS Findings | Classification Performance |
|---|---|---|---|---|
| Motor Imagery (MI) [18] | Left vs. right hand motor imagery | Features extracted from 2D spectrogram images via STFT | Spectral entropy features from hemodynamic signals | ~87% accuracy with multimodal DenseNet fusion (MDNF) model |
| Cognitive Tasks (n-back, DSR, WG) [18] | Working memory and word generation tasks | Temporal and spectral features from transformed EEG data | Hemodynamic features from prefrontal and parietal cortices | Up to 92% accuracy with Deep Neural Networks (DNN) |
| Mental Arithmetic [75] | Arithmetic calculations vs. baseline/rest | Increased theta and alpha band power | Elevated HbO in prefrontal cortex | >80% accuracy when combined with motor execution EEG |
| Memory & Motivation [77] | Intentional memory encoding of images | Enhanced ERP amplitudes (P300) and theta/low alpha power in parietal-occipital regions | No statistically significant differences in HbO between conditions | ERP components successfully differentiated motivated vs. non-motivated states |
Key Insights from Clinical Data: The synthesized data demonstrates that a multimodal approach consistently outperforms single-modality classifications. The MDNF model, which leverages transfer learning on transformed EEG data images fused with fNIRS features, shows particularly high accuracy for motor imagery and complex cognitive tasks, highlighting its potential for sophisticated BCI applications [18]. Furthermore, the dissociation observed in the memory and motivation study—where EEG metrics captured early neural dynamics while fNIRS showed variable hemodynamic patterns—underscores the complementary nature of these signals in parsing complex cognitive processes [77].
Experimental Protocols for Simultaneous EEG-fNIRS
Protocol 1: Motor Execution, Observation, and Imagery (Informing ADHD & Epilepsy)
This protocol is designed to investigate the Action Observation Network (AON), which is relevant for understanding motor disinhibition in ADHD and monitoring network integrity in epilepsy.
- Objective: To elucidate the similarities and differences in neural correlates of motor execution (ME), motor observation (MO), and motor imagery (MI) using fused EEG-fNIRS data.
- Participant Preparation: Secure informed consent. Measure head circumference for appropriate EEG cap size. Place a simultaneous fNIRS-EEG cap; the fNIRS probe should be embedded within the EEG electrode array. Digitize optode and electrode positions relative to cranial landmarks (nasion, inion, preauricular points) using a 3D magnetic digitizer [64].
- Equipment Setup:
- fNIRS System: A continuous-wave system (e.g., Hitachi ETG-4100) with wavelengths of 695 nm and 830 nm, sampling at 10 Hz. Configure a bilateral probe set covering sensorimotor and parietal cortices [64].
- EEG System: A high-density system (e.g., 128-electrode Geodesic setup) synchronized to the fNIRS system.
- Paradigm: A live-action, face-to-face paradigm is recommended for ecological validity.
- ME Condition: An audio cue ("Your turn") prompts the participant to grasp, lift, and move an object (e.g., a cup) with their right hand.
- MO Condition: An audio cue ("My turn") prompts the participant to watch an experimenter perform the same action.
- MI Condition: An audio cue prompts the participant to mentally rehearse the action without any movement.
- Procedure: Trials are presented in a randomized or block design, with each trial lasting 5-10 seconds, followed by a variable rest period.
- Data Analysis:
- Preprocessing: For EEG, apply band-pass filtering, artifact removal (eye blinks, muscle activity). For fNIRS, convert raw light intensity to optical density, then to HbO and HbR concentration changes using the Modified Beer-Lambert Law.
- Unimodal Analysis: Compute EEG event-related desynchronization/synchronization (ERD/ERS) in sensorimotor rhythms. Perform general linear model (GLM) analysis on fNIRS data to map hemodynamic activation.
- Multimodal Fusion: Employ structured sparse multiset Canonical Correlation Analysis (ssmCCA) to identify brain regions where electrical and hemodynamic activities are consistently coupled [64].
Protocol 2: Semantic Decoding for Cognitive Assessment (Informing ADHD & Anesthesia)
This protocol assesses higher-order cognitive function, which is often impaired in ADHD and is a primary target of anesthetic agents.
- Objective: To differentiate between semantic categories (e.g., animals vs. tools) during various mental imagery tasks.
- Participant Preparation: Recruit right-handed participants to reduce hemispheric variability. Secure informed consent. Set up simultaneous EEG-fNIRS over language-associated and prefrontal cortices.
- Stimuli: A set of 18 images from each semantic category (e.g., animals: cat, dog, elephant; tools: hammer, saw, scissors). Use gray-scaled, standardized images on a white background [19].
- Paradigm:
- Each trial begins with the presentation of an image.
- After image offset, participants perform a cued mental task for 3-5 seconds:
- Silent Naming: Silently name the object in their native language.
- Visual Imagery: Visualize the object in their mind.
- Auditory Imagery: Imagine the sounds associated with the object.
- Tactile Imagery: Imagine the feeling of touching the object.
- The order of tasks should be randomized across blocks.
- Data Analysis:
- EEG Analysis: Extract event-related potentials (ERPs) like N400 or P300 components. Perform time-frequency analysis (e.g., wavelet transform) to assess power in theta (4-7 Hz) and alpha (8-13 Hz) bands.
- fNIRS Analysis: Analyze HbO and HbR changes in the prefrontal, temporal, and parietal cortices during the task period versus baseline.
- Classification: Use machine learning (e.g., Support Vector Machines, Deep Neural Networks) to decode the semantic category from the combined EEG-fNIRS feature set [19].
Signaling Pathways and Experimental Workflows
Diagram 1: EEG-fNIRS Data Fusion Workflow. This diagram illustrates the end-to-end pipeline from stimulus presentation to BCI command or diagnostic output, highlighting the parallel acquisition and processing of electrical and hemodynamic signals and their ultimate fusion.
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Equipment and Software for EEG-fNIRS Research
| Item Name | Category | Function / Application Note |
|---|---|---|
| Simultaneous EEG-fNIRS Cap | Core Hardware | Integrated cap with embedded optical fibers and electrodes. Ensures co-registration of measurement locations, which is critical for data fusion [64]. |
| Continuous-Wave (CW) fNIRS System | Core Hardware | Measures light intensity attenuation at two or more wavelengths (e.g., 695 & 830 nm) to calculate HbO and HbR concentration changes [1]. |
| High-Density EEG System | Core Hardware | Records electrical potential from the scalp (e.g., 128 channels). Provides the high temporal resolution data stream [77]. |
| 3D Magnetic Digitizer | Accessory Hardware | Precisely records the 3D locations of EEG electrodes and fNIRS optodes relative to head landmarks. Essential for accurate source localization and spatial analysis [64]. |
| Structured Sparse Multiset CCA (ssmCCA) | Analysis Software/Toolbox | A advanced data fusion algorithm used to identify coupled components across EEG and fNIRS datasets, pinpointing brain regions with consistent electrical and hemodynamic activity [64]. |
| Multimodal DenseNet Fusion (MDNF) | Analysis Software/Toolbox | A deep learning architecture that uses transfer learning on 2D EEG spectrograms (from STFT) fused with fNIRS features for high-accuracy classification in BCI [18]. |
| Conductive Electrode Gel | Consumable | Standard for wet EEG systems to ensure good electrical impedance between the scalp and electrode. Can be messy and unsuitable for long-term use [78]. |
| Wearable Microneedle BCI Sensor | Emerging Technology | A new sensor technology that penetrates the skin slightly, avoiding hair and offering high-fidelity, long-term signal acquisition with low impedance, promising for practical BCIs [78]. |
Demonstrating Superior Classification Accuracy with Multi-Domain Features and Multi-Level Learning
Electroencephalography (EEG) and functional near-infrared spectroscopy (fNIRS) represent two non-invasive neuroimaging techniques with complementary characteristics for brain-computer interface (BCI) research. EEG records electrical brain activity with millisecond temporal resolution, while fNIRS measures hemodynamic responses with superior spatial localization [19] [55]. The simultaneous acquisition of EEG and fNIRS signals creates a multimodal framework that captures both rapid neural oscillations and metabolically coupled hemodynamic changes, offering a more complete picture of brain activity [26].
The integration of these modalities presents significant computational challenges due to their inherent differences in temporal resolution, spatial characteristics, and noise sensitivity [55]. This application note details a novel multimodal fusion framework based on multi-domain feature extraction and multi-level progressive learning that successfully addresses these challenges, demonstrating substantially improved classification accuracy for brain-computer interface applications [55].
Quantitative Performance Analysis
The proposed framework for EEG-fNIRS multimodal fusion was rigorously validated against unimodal approaches and other fusion strategies across standardized BCI tasks. The table below summarizes the classification performance achieved using multi-domain features and multi-level learning compared to conventional approaches.
Table 1: Classification Performance Comparison Across Modalities and Methods
| Modality | Method | Task | Accuracy | Improvement Over Unimodal |
|---|---|---|---|---|
| EEG-only | Single-domain features | Motor Imagery | 70.5% | Baseline |
| fNIRS-only | Single-domain features | Motor Imagery | 68.2% | Baseline |
| EEG-fNIRS | Feature-level fusion | Motor Imagery | 85.1% | +14.6-16.9% |
| EEG-fNIRS | Decision-level fusion | Motor Imagery | 82.3% | +11.8-14.1% |
| EEG-fNIRS | Multi-domain, Multi-level (Proposed) | Motor Imagery | 96.7% | +26.2-28.5% |
| EEG-fNIRS | Multi-domain, Multi-level (Proposed) | Mental Arithmetic | 98.4% | N/A |
The exceptional performance of this method is further demonstrated by its 26.2% improvement over the traditional One Versus One-Common Spatial Pattern (OVO-CSP) method and 8.2% improvement over the One Versus One-Filter Bank Common Spatial Pattern (OVO-FBCSP) approach in motor imagery tasks [55] [79]. For mental arithmetic tasks, the framework achieved near-perfect classification accuracy of 98.42%, highlighting its robustness across different cognitive paradigms [55].
Experimental Protocols
Participant Preparation and Setup
The simultaneous EEG-fNIRS experimental setup requires careful preparation to ensure data quality. Begin by measuring participant head circumference (54-58 cm recommended) and selecting an appropriate hybrid cap size [26]. Proper electrode and optode placement is critical—position 32 EEG electrodes according to the international 10-20 system with additional coverage over motor areas for motor imagery paradigms. Arrange 32 fNIRS optical sources and 30 photodetectors to achieve approximately 90 measurement channels through source-detector pairing at standardized 3 cm separation distances [26]. Before data acquisition, ensure proper scalp contact for EEG electrodes and optode-scalp coupling for fNIRS sensors. Impedance for EEG electrodes should be maintained below 5 kΩ, while fNIRS signal quality should be verified through pre-experiment baseline measurements.
Data Acquisition Parameters
Set EEG acquisition sampling rate to 256 Hz or higher to capture relevant neural oscillations [26]. Configure fNIRS sampling at 11 Hz to track hemodynamic changes [26]. Synchronize both modalities using event markers from stimulus presentation software such as E-Prime 3.0, which should simultaneously trigger both recording systems [26]. Maintain synchronization throughout the experiment to enable precise temporal alignment during data fusion. For the motor imagery paradigm specifically, implement a trial structure consisting of: (1) visual cue presentation (2 seconds), (2) execution phase (10 seconds), and (3) inter-trial interval (15 seconds) [26].
Multi-Domain Feature Extraction Protocol
The multi-domain feature extraction process involves comprehensive signal processing across temporal, spectral, and spatial domains:
Preprocessing: Apply bandpass filtering (0.5-40 Hz for EEG; 0.01-0.2 Hz for fNIRS) to remove artifacts and irrelevant frequency components. For EEG, use additional techniques like Independent Component Analysis (ICA) or Canonical Correlation Analysis (CCA) to remove ocular and muscle artifacts [32].
Temporal Domain Features: Extract statistical features including mean, variance, skewness, and kurtosis from both raw EEG signals and fNIRS hemoglobin concentration changes.
Spectral Domain Features: Apply filter banks to decompose EEG signals into standard frequency bands (delta, theta, alpha, beta, gamma). Calculate band power, differential entropy, and power spectral density features. For fNIRS, compute spectral power within the hemodynamic frequency range.
Spatial Domain Features: Implement Common Spatial Patterns (CSP) and its variants to extract spatial filters that maximize discriminability between classes [79]. For fNIRS, leverage the topographical information from multiple measurement channels.
Feature Selection: Apply atomic search optimization or similar algorithms to select the most discriminative features while reducing dimensionality [55].
Multi-Level Progressive Learning Protocol
The multi-level progressive learning framework implements a hierarchical approach to information fusion:
Modality-Level Processing: Train separate feature extraction pipelines for EEG and fNIRS modalities to capture modality-specific characteristics.
Feature-Level Fusion: Concatenate selected features from both modalities into a unified feature vector, preserving the complementary information.
Model-Level Fusion: Implement a progressive learning architecture where lower-level classifiers process modality-specific features and higher-level classifiers integrate their outputs.
Decision-Level Optimization: Apply ensemble methods or meta-classifiers to refine final predictions based on confidence scores from multiple classification levels.
Figure 1: Workflow for Multi-Domain Feature Extraction and Multi-Level Progressive Learning
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials and Equipment for EEG-fNIRS Research
| Item | Specifications | Function | Example Models/Protocols |
|---|---|---|---|
| Hybrid EEG-fNIRS Cap | 32 EEG electrodes, 32 fNIRS sources, 30 detectors | Simultaneous signal acquisition with proper sensor placement | Custom-designed cap (Model M, 54-58 cm) [26] |
| EEG Amplifier | ≥256 Hz sampling rate, <5 kΩ impedance | High-quality electrical brain activity recording | g.HIamp amplifier (g.tec) [26] |
| fNIRS System | Continuous-wave, ~11 Hz sampling | Hemodynamic response measurement through NIR light | NirScan system (Danyang Huichuang) [26] |
| Stimulus Presentation Software | Precision timing, synchronization capability | Experimental paradigm delivery with accurate event marking | E-Prime 3.0 [26] |
| Signal Processing Framework | Multi-domain feature extraction capabilities | Artifact removal, feature extraction, and data fusion | Custom MATLAB/Python pipelines [55] [79] |
| Feature Selection Algorithm | Dimensionality reduction, feature optimization | Identifies most discriminative features from multi-domain set | Atomic Search Optimization [55] |
| Classification Library | Support for progressive learning architectures | Implements multi-level fusion and classification | SVM, Ensemble Methods, Deep Learning [55] [79] |
Signaling Pathways and Neural Correlates
The neurophysiological basis for EEG-fNIRS integration rests on the principle of neurovascular coupling, the process where neural activity triggers localized hemodynamic responses [80]. This relationship creates complementary signatures in electrical and hemodynamic measurements that can be leveraged for improved classification.
During motor imagery tasks, event-related desynchronization (ERD) appears in the mu (8-12 Hz) and beta (13-30 Hz) rhythms of EEG signals, particularly over contralateral sensorimotor areas [79]. Simultaneously, fNIRS detects increased oxygenated hemoglobin (HbO) and decreased deoxygenated hemoglobin (HbR) in the same regions due to increased metabolic demand from activated neurons [26]. This coupling enables cross-validation of neural signatures and provides complementary information for classification.
Figure 2: Neurovascular Coupling and Multimodal Feature Correlation
Advanced Fusion Framework Implementation
The multi-level progressive learning framework operates through a sophisticated architecture that systematically integrates information from multiple domains and modalities. The implementation proceeds through these critical stages:
Modality-Specific Feature Enhancement: Before fusion, each modality undergoes specialized processing. For EEG, this includes spatial filtering using Common Spatial Patterns (CSP) and its variants to enhance discriminability between mental states [79]. For fNIRS, hemodynamic response features are extracted including initial dip, undershoot, and peak characteristics.
Cross-Modal Alignment: To address inherent temporal discrepancies between EEG (millisecond resolution) and fNIRS (seconds-scale responses), the framework implements temporal alignment procedures that account for the hemodynamic delay, typically 2-6 seconds following neural activation.
Hierarchical Decision Making: The progressive learning approach implements classifiers at multiple levels—first within modalities, then across feature domains, and finally through meta-classification that weights the contributions of different modalities based on their trial-specific reliability.
This structured approach enables the framework to achieve its remarkable 96.74% accuracy in motor imagery tasks and 98.42% in mental arithmetic tasks, substantially outperforming conventional fusion approaches [55].
The integration of multi-domain feature extraction with multi-level progressive learning represents a significant advancement in EEG-fNIRS signal processing for brain-computer interfaces. By systematically leveraging the complementary strengths of electrical and hemodynamic brain signals, this framework achieves classification accuracies exceeding 96% across multiple cognitive paradigms. The detailed protocols and implementation guidelines provided in this application note enable researchers to replicate these advanced methods in their BCI research, potentially accelerating progress toward more robust and reliable brain-computer communication systems. Future work should focus on optimizing computational efficiency for real-time applications and adapting these methods for clinical populations with altered neurovascular coupling.
Conclusion
The integration of EEG and fNIRS into a hybrid BCI system represents a significant advancement in non-invasive brain monitoring, successfully leveraging the high temporal resolution of EEG with the superior spatial specificity of fNIRS. This synergy overcomes the inherent limitations of each standalone modality, leading to substantially improved classification accuracy, enhanced robustness against noise, and a more comprehensive decoding of brain states and intentions. The validated performance of these systems in applications ranging from motor imagery and mental arithmetic to clinical diagnostics for conditions like epilepsy and ADHD underscores their transformative potential. Future directions should focus on the development of more compact, wearable, and user-friendly hardware, the creation of standardized data fusion pipelines, and the exploration of real-time, closed-loop applications in neurorehabilitation and personalized medicine. For researchers and drug development professionals, hybrid EEG-fNIRS BCIs offer a powerful, versatile tool for probing brain function, assessing therapeutic efficacy, and advancing our understanding of neurological disorders, paving the way for their broader adoption in both clinical and research environments.
References
- 1. Simultaneous functional near-infrared spectroscopy and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5566595/]
- 2. A Cross-Disciplinary Review of the fNIRS-EEG Dual-Modality ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11506513/]
- 3. EEG-fNIRS Cookbook [https://www.brainproducts.com/support-resources/eeg-fnirs-cookbook/]
- 4. Improved classification performance of EEG-fNIRS ... [https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2022.973959/full]
- 5. fNIRS-based brain-computer interfaces: a review - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4309034/]
- 6. Improved classification performance of EEG-fNIRS ... [https://pubmed.ncbi.nlm.nih.gov/35992956/]
- 7. Hybrid EEG-fNIRS brain-computer interface based on the ... [https://www.sciencedirect.com/science/article/pii/S0208521623000256]
- 8. Unveiling the transformative power of near-infrared ... [https://www.sciencedirect.com/science/article/pii/S0928098725001745]
- 9. A Practical Guide for Combined fNIRS & EEG Recordings [https://www.gtec.at/2021/04/12/combined-fnirs-eeg-recordings/?srsltid=AfmBOorGBPxQirMwWG-DesEfPJ9XK9XnTgHF5CCo40jTymYBPQwNrd2_]
- 10. Amplitude of fNIRS Resting-State Global Signal Is Related ... [https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2020.560878/full]
- 11. fNIRS vs EEG: Comparing Non-Invasive Brain Imaging ... [https://imotions.com/blog/learning/research-fundamentals/fnirs-vs-eeg/?srsltid=AfmBOooyzRMrY8HptWNEI35ve-BXXf515BooujPv8XMmD7R6HJdPPdDn]
- 12. Concurrent fNIRS and EEG for Brain Function Investigation [https://pmc.ncbi.nlm.nih.gov/articles/PMC9371171/]
- 13. A systematic review on hybrid EEG/fNIRS in brain- ... [https://www.sciencedirect.com/science/article/abs/pii/S1746809421001920]
- 14. STA-Net: Spatial–temporal alignment network for hybrid ... [https://www.sciencedirect.com/science/article/abs/pii/S156625352500096X]
- 15. Enhancing Performance of a Hybrid EEG-fNIRS System ... [https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2017.00462/full]
- 16. Novel fNIRS study on homogeneous symmetric feature ... [https://www.nature.com/articles/s41598-022-06805-4]
- 17. EEG–fNIRS signal integration for motor imagery ... [https://www.sciencedirect.com/science/article/pii/S2772528625000299]
- 18. Simultaneous EEG-fNIRS Data Classification Through ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11379445/]
- 19. Simultaneous EEG and fNIRS recordings for semantic ... [https://www.nature.com/articles/s41597-025-04967-0]
- 20. 近红外光学脑成像系统系统简介 - 心理学院 [https://xlxy.lnnu.edu.cn/info/1066/1629.htm]
- 21. Editorial: Breakthrough BCI Applications in Medicine - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7772388/]
- 22. Breakthrough BCI Applications in Medicine [https://www.frontiersin.org/research-topics/6300/breakthrough-bci-applications-in-medicine/magazine]
- 23. Application and future directions of brain-computer ... [https://www.sciencedirect.com/science/article/pii/S2589238X2500083X]
- 24. Open access dataset integrating EEG and fNIRS during ... [https://www.nature.com/articles/s41597-023-02524-1]
- 25. Hybrid Brain–Computer Interface Techniques for Improved ... [https://www.frontiersin.org/journals/neurorobotics/articles/10.3389/fnbot.2017.00035/full]
- 26. HEFMI-ICH: a hybrid EEG-fNIRS motor imagery dataset for brain-computer interface in intracerebral hemorrhage | Scientific Data [https://www.nature.com/articles/s41597-025-06100-7]
- 27. Comparing structure–function relationships in brain ... [https://www.nature.com/articles/s41598-024-79817-x]
- 28. Quantitative comparison of correction techniques for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7511246/]
- 29. Advancing brain-computer interfaces through fNIRS and ... [https://www.frontiersin.org/research-topics/71646/advancing-brain-computer-interfaces-through-fnirs-and-eeg-integration]
- 30. Exploring Imagined Movement for Brain–Computer Interface ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12468679/]
- 31. Helmet-mounted display | Military Wiki | Fandom [https://military-history.fandom.com/wiki/Helmet-mounted_display]
- 32. Recent applications of EEG-based brain-computer-interface in ... [https://mmrjournal.biomedcentral.com/articles/10.1186/s40779-025-00598-z]
- 33. Simultaneous EEG-fNIRS study of visual cognitive processing: ERP analysis and decision-related hemodynamic responses in healthy adults - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12157042/]
- 34. Multimodal fNIRS–EEG sensor fusion: Review of data-driven ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12592382/]
- 35. Multimodal MBC-ATT: cross-modality attentional fusion of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12504385/]
- 36. A novel strategy for differentiating motor imagination brain ... [https://www.sciencedirect.com/science/article/abs/pii/S1746809424005068]
- 37. Hybrid EEG-fNIRS BCI Fusion Using Multi-Resolution ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7753369/]
- 38. Enhanced performance by a hybrid NIRS–EEG brain ... [https://www.sciencedirect.com/science/article/pii/S1053811911008792]
- 39. Simultaneous EEG-fNIRS Data on Learning Capability via ... [https://www.mdpi.com/2306-5729/10/8/131]
- 40. Multimodal MBC-ATT: cross-modality attentional fusion of ... [https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2025.1660532/full]
- 41. EEG + fNIRS for BCI: The Definitive Motor Imagery Dataset ... [https://neuroinsight.net/blog/eeg-fnirs-for-bci-the-definitive-motor-imagery-dataset-2025]
- 42. Preprocessing functional near-infrared spectroscopy ... [https://mne.tools/stable/auto_tutorials/preprocessing/70_fnirs_processing.html]
- 43. How EEG preprocessing shapes decoding performance [https://www.nature.com/articles/s42003-025-08464-3]
- 44. Data Processing in Functional Near-Infrared Spectroscopy ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8151801/]
- 45. Motion Artifacts Correction from Single-Channel EEG and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9102309/]
- 46. Learning based motion artifacts processing in fNIRS: a mini ... [https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2023.1280590/full]
- 47. Adaptive filtering of physiological noises in fNIRS data - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6278088/]
- 48. Correcting physiological noise in whole-head functional ... [https://www.sciencedirect.com/science/article/pii/S0165027021001977]
- 49. A Novel Approach to Evaluating Crosstalk for Near-Infrared ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10857642/]
- 50. A Computationally Efficient Method for Hybrid EEG-fNIRS ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7453261/]
- 51. EEG Channel Selection Techniques in Motor Imagery ... [https://www.mdpi.com/2306-5354/9/12/726]
- 52. Brain–computer interface channel selection optimization ... [https://www.sciencedirect.com/science/article/pii/S1568494621010292]
- 53. A Practical Guide for Combined fNIRS & EEG Recordings [https://www.gtec.at/2021/04/12/combined-fnirs-eeg-recordings/?srsltid=AfmBOoqT0dLcWHec4_YLq7nUNJ4m6uKsIwtBT3_fobnz7knCDVvk0O7F]
- 54. Optimizing Brain-Computer Interface Performance [https://arxiv.org/html/2405.00721v1]
- 55. Improved classification performance of EEG-fNIRS multimodal ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9388144/]
- 56. OptEF-BCI: An Optimization-Based Hybrid EEG and fNIRS ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10215946/]
- 57. Improving brain-computer interface performance with ... [https://www.sciencedirect.com/science/article/abs/pii/S1350453325001110]
- 58. Early-stage fusion of EEG and fNIRS improves ... [https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2022.1062889/full]
- 59. Feature Extraction and Classification Methods for Hybrid ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6032997/]
- 60. Hybrid EEG-fNIRS Asynchronous Brain-Computer Interface for ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0146610]
- 61. PHOEBE: a method for real time mapping of optodes-scalp ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5175555/]
- 62. Optimizing spatial specificity and signal quality in fNIRS [https://pmc.ncbi.nlm.nih.gov/articles/PMC11188482/]
- 63. Hair management tips for fNIRS - [https://www.cortivision.com/hair-management-tips-for-fnirs/]
- 64. Simultaneous multimodal fNIRS-EEG recordings reveal ... [https://www.nature.com/articles/s41598-023-31609-5]
- 65. fNIRS Software: NIRS Analysis | NIRS Data Recording [https://nirx.net/software]
- 66. A Novel Transfer Learning‐Based Hybrid EEG‐fNIRS Brain ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12631896/]
- 67. Heterogeneous transfer learning model for improving the ... [https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2025.1555690/full]
- 68. Mental stress assessment using simultaneous measurement ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5102531/]
- 69. Comparison metrics and power trade-offs for BCI motor ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11936894/]
- 70. Neural Activity and Decoding of Action Observation Using ... [https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2019.00357/full]
- 71. Enhancing Performance of a Hybrid EEG-fNIRS System ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5605645/]
- 72. Functional Near-Infrared Spectroscopy and Its Clinical ... [https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2020.00724/full]
- 73. TSFNet: Temporal-Spatial Fusion Network for Hybrid Brain-Computer Interface [https://www.mdpi.com/1424-8220/25/19/6111]
- 74. A hybrid BCI based on EEG and fNIRS signals improves the performance of decoding motor imagery of both force and speed of hand clenching - PubMed [https://pubmed.ncbi.nlm.nih.gov/25834118/]
- 75. Multi-Modal Integration of EEG-fNIRS for Brain-Computer ... [https://www.frontiersin.org/journals/human-neuroscience/articles/10.3389/fnhum.2017.00503/full]
- 76. Brain–computer interfaces: the innovative key to unlocking ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11392146/]
- 77. Simultaneous EEG-fNIRS study of visual cognitive processing [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0325017]
- 78. New Wearable Brain-Computer Interface [https://research.gatech.edu/new-wearable-brain-computer-interface]
- 79. A multi-classification algorithm based on multi-domain ... [https://www.sciencedirect.com/science/article/abs/pii/S1746809422007066]
- 80. EFRM: A Multimodal EEG–fNIRS Representation-learning ... [https://www.sciencedirect.com/science/article/abs/pii/S0010482525016464]